Biology:Snf3
| High-affinity glucose transporter SNF3 | |
|---|---|
| Identifiers | |
| Symbol | SNF3 |
| RefSeq | NP_010087.1 |
| UniProt | P10870 |
Snf3 is a protein which regulates glucose uptake in yeast. It senses glucose in the environment with high affinity.
Introduction
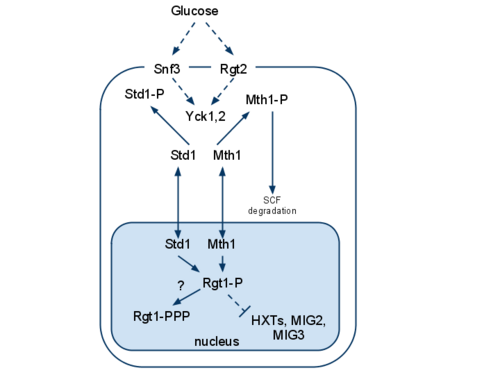
Glucose sensing and signaling in budding yeast is similar to the mammalian system in many ways. However, there are also significant differences. Mammalian cells regulate their glucose uptake via hormones (i.e. insulin and glucagon) or intermediary metabolites. In contrast, yeast as a unicellular organism does not depend on hormones but on nutrients in the medium. The presence of glucose induces a conformational change in the membrane proteins Snf3/Rgt2 or Gpr1, and regulates expression of genes involved in glucose metabolism.
Homology and function
Snf3 is homologous to multiple sugar transporters, it shares high similarity to the glucose transporters of rat brain cells and human HepG2 hepatoma cells, as well as to the arabinose and xylose transporters (AraE and XylE) of Escherichia coli.[1] Based on this homology and on genetic studies, Snf3 was initially thought to be a high affinity glucose transporter.
Later, it was found that Snf3 is not a glucose transporter, but rather a high affinity glucose sensor. It senses glucose at low concentrations and regulates transcription of the HXT genes, which encode for glucose transporters. If glucose is absent Snf3 is quiescent and transcription of the HXT genes is inhibited by a repressing complex. The complex consisting of several subunits such as Rgt1, Mth1/Std1, Cyc8 and Tup1 binds to the promoters of the HXT genes, thereby blocking their transcription.

Snf3 is able to bind even low amounts of glucose due to its high affinity. The induction of Snf3 by glucose leads to the activation of YckI, a yeast casein kinase. This is followed by the recruitment of Mth1 and Std1 to the C-terminus of Snf3 which facilitates the phosphorylation of the two proteins by YckI. Phosphorylated Mth1 and Std1 are subsequently tagged for proteasome dependent degradation by SCFGrrl, an E3 ubiquitin ligase. Therefore, the inhibitory complex misses two of its key components and cannot be assembled. Thus, repression of the HXT genes is abolished, leading to the expression of the glucose transporters and subsequently glucose import.[2]
Structure
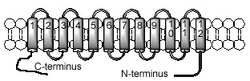
Snf3 is a plasma membrane protein in yeasts that consists of 12 (2x6) transmembrane domains, like the homologous glucose transporters. Its structure is distinct from the homologous transporters in particular by a long C-terminal tail which is predicted to reside in the cytoplasm.
The long C-terminal tail plays an important role in glucose signaling and is probably the signaling domain itself. A soluble version of the C-terminal tail alone is sufficient to induce glucose transport.[1][2][3]
All glucose transporters including Snf3 contain an arginine residue situated in a cytoplasmic loop preceding the fifth transmembrane domain. If this position is mutated, Snf3 adopts a state of constant glucose induction irrespective of whether there are nutrients present or not; this suggests an involvement in the glucose sensing process.
Regulation
The regulation of Snf3 in S. cerevisiae and its downstream events are still poorly understood, but it seems clear that a second glucose sensor Rgt2 influences Snf3 and vice versa. Furthermore, it is unclear whether these two proteins sense the glucose concentration on the outside or inside the cell. Snf3 and Rgt2 influence directly or indirectly several Hxt-transporters which are responsible for the glucose uptake. Low extracellular glucose concentrations are sensed by the Snf3 protein which probably leads to the expression of Hxt2-Genes for high affinity glucose transporters, while Rgt2 senses high glucose concentrations and leads to the expression of low affinity glucose transporters, like Hxt1 Although the downstream pathway is poorly understood it seems that Snf3 and Rgt2 transmit a signal directly or indirectly to Grr1, the DNA binding protein Rgt1, and the two cofactors Ssn6 and Tup1. Also needed for the transcription are the two nuclear proteins Mth1 and Std1.[4]
References
- ↑ 1.0 1.1 Celenza JL, Marshall-Carlson L, Carlson M (1988). The yeast SNF3 gene encodes a glucose transporter homologous to the mammalian protein. PNAS 85, 2130-2134
- ↑ 2.0 2.1 Rolland F, Winderickx J, Thevelein JM. (2002). Glucose-sensing and -signaling mechanisms in yeast. FEMS Yeast Res. 2, 183-201
- ↑ Marshall-Carlson L, Celenza JL, Laurent BC, Carlson M (1990). Mutational analysis of the SNF3 Glucose Transporter of Saccharomyces cerevisiae. Mol Cell Biol 10(3), 1105-1115
- ↑ Schneper L, Düvel K, Broach JR (2004). Sense and sensibility: nutritional response and signal integration in yeast. Current Opinion in Microbiology 7, 624-630
- Gancedo MJ (2008). The early steps of glucose signaling in yeast. FEMS Microbiol Rev 32, 673-704
- Kruckeberg AL, Walsh MC, Van Dam K (1998). How do yeast cells sense glucose? BioEssays 20, 972-976
- Schneper L, Düvel K, Broach JR (2004). Sense and sensibility: nutritional response and signal integration in yeast. Current Opinion in Microbiology 7, 624-630
 |
