Biology:WDR90
WD repeat-containing protein 90 is a protein that, in humans, is encoded by the WDR90 gene (16p13.3).[1] This human protein is 1750 amino acids, and has a molecular weight of 187.7 kDa.[2] It contains multiple WD40 repeat domains and one domain of unknown function.[3] This protein is conserved all the way back to invertebrates.[4] Proteins containing WD transducin repeating domains have been found to play a role in a variety of functions ranging from signal transduction and transcription regulation to cell cycle control, autophagy and apoptosis.[5]
Gene
In the human genome, WDR90 spans from base pairs 649363 to 667833, and has an mRNA sequence that is 5,250 nucleotides in length. This gene consists of 41 exons and encodes for a 1,750 amino acid protein in humans.
Locus
This human gene is located in the positive DNA strand on chromosome 16 at the position 16p13.3.[1] Flanking sequences on the positive strand include FAM195A and RHOT2.[3]
Aliases
Aliases for WDR90 include: C16orf15, C16orf16, C16orf17, C16orf18, C16orf19, and KIAA1924.[1]
Homology
Paralogs
One human paralog for WDR90 has been found. This paralog is known as WDR16. According to UniProtKB/Swiss-Prot, WDR16 may play an essential role in growth or survival of hepatocellular carcinoma. This protein in humans if 620 amino acids long.[6][7]
Orthologs
Orthologs for protein WDR90 are found conserved all the way back to single invertebrates.[8] There were three conserved domains across mammals, DUF667 (location 3-124), and two WD40 repeats (locations 697-991, and 1155-1549).[3]
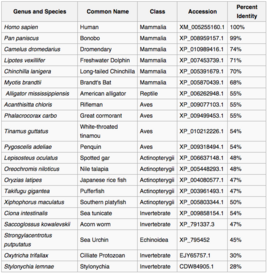
Protein
Primary sequence
Human protein WDR90 is 287.7 kDa, with an isoelectric point of 6.584. There is a domain of unknown function called DUF667 that is conserved across its 30 predicted isoforms.[9]
Post-translational modifications
WDR90 is predicted to be a heavily phosphorylated protein.[10]
Domains and motifs
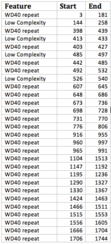
There are three domains that repeat throughout the human sequence.[11]
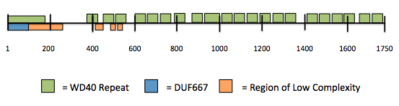
Secondary structure
The de novo protein structure of WDR90 in humans is predicted to possess beta sheets, beta turns, and random coils.[12]
Tertiary and quaternary structure
The human protein WDR90 adopts a 7-bladed beta propeller that is indicative of the WD40 repeat family. This structure was predicted with 100% confidence by the structure predicting program Phyre2.
Interacting proteins
The most notable proteins that interact with human WDR90 are predicted to be UTY, KDM6B, SETD1B, and ASH2L.[13] These proteins are primarily found as histone modifiers.
Transcription factors
The promoter for WDR90 in humans has been predicted to have domains for the transcription factors: ARNT, AP-1, MYB, and Cis2His2.[14]
Expression
Expression levels of human WDR90 appear to be relatively constituent in low levels, with its highest levels in the lymph nodes and thymus.[15][16] The expression of WDR90 in development is the highest in blastocyst and fetal stages. Expression is notably absent from the neonatal and infant stages, then comes back during juvenile and persists through adulthood.[16]
Function
The function of WDR90 in humans in not well known. Given its broad expression in many tissues, its ties to the protein superfamily WD40 domain-containing proteins, and the known protein interactions, it could play a role in histone modifications, vesicular transport, or transcription regulation.[13][15][16]
References
- ↑ 1.0 1.1 1.2 Database, GeneCards. "WDR90 Gene - GeneCards | WDR90 Protein | WDR90 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=WDR90. Retrieved 2015-05-06.
- ↑ Volker Brendel, Department of Mathematics, Stanford University, Stanford CA 94305, U.S.A., modified; any errors are due to the modification. http://workbench.sdsc.edu/
- ↑ 3.0 3.1 3.2 "WDR90 WD repeat domain 90 [Homo sapiens (Human)] - Gene - NCBI". https://www.ncbi.nlm.nih.gov/gene/197335.
- ↑ "NCBI - WWW Error Blocked Diagnostic". https://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?UGID=902650&TAXID=9606&SEARCH=WDR90. Retrieved 2015-04-30.
- ↑ Stirnimann, Christian U.; Petsalaki, Evangelia; Russell, Robert B.; Müller, Christoph W. (Oct 2010). "WD40 proteins propel cellular networks". Trends in Biochemical Sciences 35 (10): 565–574. doi:10.1016/j.tibs.2010.04.003. ISSN 0968-0004. PMID 20451393.
- ↑ Database, GeneCards. "WDR16 Gene - GeneCards | WDR16 Protein | WDR16 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=WDR16&search=f93edcafc7b414b0d4fe4d1911579457. Retrieved 2015-05-06.
- ↑ "ExPASy: SIB Bioinformatics Resource Portal - Categories". http://www.expasy.org/proteomics. Retrieved 2015-05-06.
- ↑ "CLUSTALWPROF". SDSC. http://seqtool.sdsc.edu/CGI/BW.cgi#!.[yes|permanent dead link|dead link}}]
- ↑ "Biology WorkBench". Volker Brendel, Department of Mathematics, Stanford University, Stanford CA 94305, U.S.A., modified; any errors are due to the modification.. http://seqtool.sdsc.edu/CGI/BW.cgi#!.[yes|permanent dead link|dead link}}]
- ↑ "NetPhos 2.0 Server". http://www.cbs.dtu.dk/services/NetPhos/. Retrieved 2015-05-07.
- ↑ "Ensembl genome browser 79: Homo sapiens - Domains & features - Transcript: WDR90-002 (ENST00000293879)". http://useast.ensembl.org/Homo_sapiens/Transcript/Domains?db=core;g=ENSG00000161996;r=16:649311-667833;t=ENST00000293879. Retrieved 2015-05-06.
- ↑ UCBL, Institut. "NPS@ : SOPMA secondary structure prediction". https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html. Retrieved 2015-05-07.
- ↑ 13.0 13.1 "STRING: functional protein association networks". http://string-db.org/newstring_cgi/show_input_page.pl. Retrieved 2015-05-07.
- ↑ "Genomatix: Retrieve and analyze promoters: Query Input". https://www.genomatix.de/cgi-bin//c2p/c2p.pl?s=33e03e12197d2119c8cf1b5e0dc2b575;TASK=c2p;SHOW=WDR90.html. Retrieved 2015-05-07.
- ↑ 15.0 15.1 "NCBI - WWW Error Blocked Diagnostic". https://www.ncbi.nlm.nih.gov/geoprofiles. Retrieved 2015-05-07.
- ↑ 16.0 16.1 16.2 "NCBI - WWW Error Blocked Diagnostic". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.511903. Retrieved 2015-05-07.
Suggested reading
- Bayne, Rosemary AL, et al. "Molecular Profiling of the Human Testis Reveals Stringent Pathway‐Specific Regulation of RNA Expression Following Gonadotropin Suppression and Progestogen Treatment." Journal of andrology29.4 (2008): 389-403.
- Cox, D. M., and M. G. Butler. "Distal Partial Trisomy 15q26 and Partial Monosomy 16p13. 3 in a 36-Year-Old Male with Clinical Features of Both Chromosomal Abnormalities." Cytogenetic and genome research (2015).
- Huang, Chi-Cheng, et al. "Concurrent gene signatures for han chinese breast cancers." PLoS ONE 8.10 (2013): e76421.
- Jakobsen, Lis, et al. "Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods." The EMBO Journal 30.8 (2011): 1520-1535.
- Lauwerys, Bernard, et al. "Method for the determination and the classification of rheumatic conditions." U.S. Patent Application 12/528,615.
- Rink, Lori, et al. "Gene expression signatures and response to imatinib mesylate in gastrointestinal stromal tumor." Molecular Cancer Therapeutics 8.8 (2009): 2172-2182.
- Yanagisawa, Haru-aki, et al. "Station, Texas 77843, USA. Abbreviations: DMT, doublet microtubule; IJ, inner junction."
 |
