De Bruijn graph
In graph theory, an n-dimensional De Bruijn graph of m symbols is a directed graph representing overlaps between sequences of symbols. It has mn vertices, consisting of all possible length-n sequences of the given symbols; the same symbol may appear multiple times in a sequence. For a set of m symbols S = {s1, …, sm}, the set of vertices is:
If one of the vertices can be expressed as another vertex by shifting all its symbols by one place to the left and adding a new symbol at the end of this vertex, then the latter has a directed edge to the former vertex. Thus the set of arcs (that is, directed edges) is
Although De Bruijn graphs are named after Nicolaas Govert de Bruijn, they were invented independently by both de Bruijn[1] and I. J. Good.[2] Much earlier, Camille Flye Sainte-Marie[3] implicitly used their properties.
Properties
- If n = 1, then the condition for any two vertices forming an edge holds vacuously, and hence all the vertices are connected, forming a total of m2 edges.
- Each vertex has exactly m incoming and m outgoing edges.
- Each n-dimensional De Bruijn graph is the line digraph of the (n − 1)-dimensional De Bruijn graph with the same set of symbols.[4]
- Each De Bruijn graph is Eulerian and Hamiltonian. The Euler cycles and Hamiltonian cycles of these graphs (equivalent to each other via the line graph construction) are De Bruijn sequences.
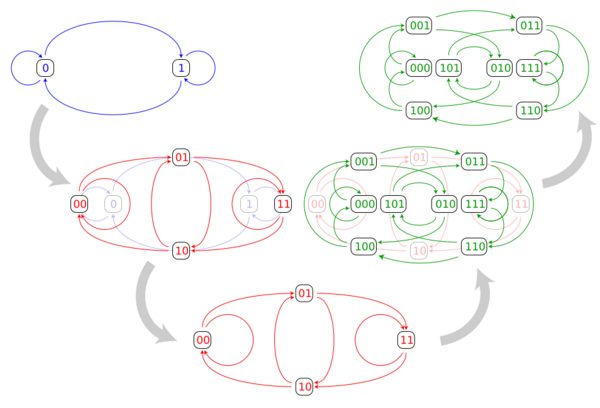
The line graph construction of the three smallest binary De Bruijn graphs is depicted below. As can be seen in the illustration, each vertex of the n-dimensional De Bruijn graph corresponds to an edge of the (n − 1)-dimensional De Bruijn graph, and each edge in the n-dimensional De Bruijn graph corresponds to a two-edge path in the (n − 1)-dimensional De Bruijn graph.
Dynamical systems
Binary De Bruijn graphs can be drawn in such a way that they resemble objects from the theory of dynamical systems, such as the Lorenz attractor: Lua error in Module:Multiple_image at line 163: attempt to perform arithmetic on local 'totalwidth' (a nil value). This analogy can be made rigorous: the n-dimensional m-symbol De Bruijn graph is a model of the Bernoulli map
The Bernoulli map (also called the 2x mod 1 map for m = 2) is an ergodic dynamical system, which can be understood to be a single shift of a m-adic number.[5] The trajectories of this dynamical system correspond to walks in the De Bruijn graph, where the correspondence is given by mapping each real x in the interval [0,1) to the vertex corresponding to the first n digits in the base-m representation of x. Equivalently, walks in the De Bruijn graph correspond to trajectories in a one-sided subshift of finite type.

Embeddings resembling this one can be used to show that the binary De Bruijn graphs have queue number 2[6] and that they have book thickness at most 5.[7]
Uses
- Some grid network topologies are De Bruijn graphs.
- The distributed hash table protocol Koorde uses a De Bruijn graph.
- In bioinformatics, De Bruijn graphs are used for de novo assembly of sequencing reads into a genome.[8][9][10][11][12] Instead of the complete De Bruijn graphs described above that contain all possible k-mers, de novo sequence assemblers make use of De Bruijn subgraphs that contain only the k-mers observed in a sequencing dataset.
- In time series forecasting, De Bruijn graphs have been adapted to encode temporal patterns by mapping discrete subsequences (n-grams) of observations to graph nodes. This enables the modeling of sequential dependencies in symbolic or discretized time series data.[13][14] Multivariate De Bruijn graphs extend this idea by jointly encoding patterns across multiple correlated variables, allowing for the representation of complex inter-variable temporal dynamics in multivariate time series.[15]
See also
References
- ↑ "A combinatorial problem". Indagationes Mathematicae 49: 758–764. 1946.
- ↑ Good, I. J. (1946). "Normal recurring decimals". Journal of the London Mathematical Society 21 (3): 167–169. doi:10.1112/jlms/s1-21.3.167.
- ↑ Flye Sainte-Marie, C. (1894). "Question 48". L'Intermédiaire des Mathématiciens 1: 107–110.
- ↑ Zhang, Fu Ji; Lin, Guo Ning (1987). "On the de Bruijn–Good graphs". Acta Mathematica Sinica 30 (2): 195–205.
- ↑ Leroux, Philippe (2005). "Coassociative grammar, periodic orbits, and quantum random walk over ". International Journal of Mathematics and Mathematical Sciences 2005 (24): 3979–3996. doi:10.1155/IJMMS.2005.3979.
- ↑ Heath, Lenwood S. (1992). "Laying out graphs using queues". SIAM Journal on Computing 21 (5): 927–958. doi:10.1137/0221055.
- ↑ Obrenić, Bojana (1993). "Embedding de Bruijn and shuffle-exchange graphs in five pages". SIAM Journal on Discrete Mathematics 6 (4): 642–654. doi:10.1137/0406049.
- ↑ "An Eulerian path approach to DNA fragment assembly". Proceedings of the National Academy of Sciences 98 (17): 9748–9753. 2001. doi:10.1073/pnas.171285098. PMID 11504945. Bibcode: 2001PNAS...98.9748P.
- ↑ "Fragment Assembly with Double-Barreled Data". Bioinformatics 17 (Suppl 1): S225–S233. 2001. doi:10.1093/bioinformatics/17.suppl_1.S225. PMID 11473013.
- ↑ Zerbino, Daniel R.; Birney, Ewan (2008). "Velvet: algorithms for de novo short read assembly using de Bruijn graphs". Genome Research 18 (5): 821–829. doi:10.1101/gr.074492.107. PMID 18349386.
- ↑ Chikhi, R.; Limasset, A.; Jackman, S.; Simpson, J.; Medvedev, P. (2014). "On the representation of de Bruijn graphs". Journal of Computational Biology 22 (5): 336–52. doi:10.1089/cmb.2014.0160. PMID 25629448.
- ↑ Iqbal, Zamin; Caccamo, Mario; Turner, Isaac; Flicek, Paul; McVean, Gil (2012). "De novo assembly and genotyping of variants using colored de Bruijn graphs". Nature Genetics 44 (2): 226–32. doi:10.1038/ng.1028. PMID 22231483.
- ↑ Cakiroglu, Mert Onur; Kurban, Hasan; Buxton, Elham Khorasani; Dalkilic, Mehmet (2024). "A Novel Discrete Time Series Representation with De Bruijn Graphs for Enhanced Forecasting Using TimesNet (Extended Abstract)". Proceedings of the 2024 IEEE 11th International Conference on Data Science and Advanced Analytics (DSAA). pp. 1–3. doi:10.1109/DSAA61799.2024.10722826.
- ↑ Cakiroglu, Mert Onur; Kurban, Hasan; Aljihmani, Lilia; Qaraqe, Khalid; Petrovski, Goran; Dalkilic, Mehmet M. (2024). "A reinforcement learning approach to effective forecasting of pediatric hypoglycemia in diabetes I patients using an extended de Bruijn graph". Scientific Reports 14 (1): 31251. doi:10.1038/s41598-024-82649-4. PMID 39732907. PMC 11682413. Bibcode: 2024NatSR..1431251C. https://doi.org/10.1038/s41598-024-82649-4.
- ↑
Mert Onur Cakiroglu, Idil Bilge Altun, Hasan Kurban, Elham Buxton, Mehmet Dalkilic (2025). "Multivariate de Bruijn Graphs: A Symbolic Graph Framework for Time Series Forecasting". arXiv:2505.22768 [cs.LG].
{{cite arXiv}}: CS1 maint: multiple names: authors list (link)
External links
- Tutorial on using De Bruijn Graphs in Bioinformatics by Homolog.us
- De Bruijn Graph algorithm tutorial by Kurban Intelligence Lab
 |