Physics:Matrix-assisted laser desorption electrospray ionization
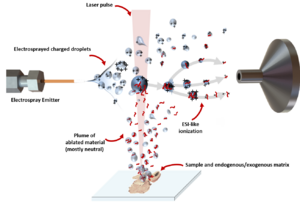
Matrix-assisted laser desorption electrospray ionization (MALDESI) was first introduced in 2006 as a novel ambient ionization technique which combines the benefits of electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI).[1] An infrared (IR) or ultraviolet (UV) laser can be utilized in MALDESI to resonantly excite an endogenous or exogenous matrix. The term 'matrix' refers to any molecule that is present in large excess and absorbs the energy of the laser, thus facilitating desorption of analyte molecules. The original MALDESI design was implemented using common organic matrices, similar to those used in MALDI, along with a UV laser. The current MALDESI source employs endogenous water or a thin layer of exogenously deposited ice as the energy-absorbing matrix where O-H symmetric and asymmetric stretching bonds are resonantly excited by a mid-IR laser.[2]
The IR-MALDESI source can be used for mass spectrometry imaging (MSI), a technique using MS data analyzed from a sample area to detect hundreds to thousands of biomolecules and visualize their spatial distributions. The IR-MALDESI MSI source was designed and implemented in 2013,[3] and is coupled to a high resolving power hybrid Quadrupole-Orbitrap mass spectrometer. The computer controlled motorized stage and a charge-coupled device (CCD) camera are placed in a nitrogen purged enclosure where ambient ions and relative humidity can be regulated. A water-cooled Peltier thermoelectric plate is used to control the sample stage temperature (−10 °C to 80 °C). The source has single- or multi-shot capabilities with adjustable laser fluence, repetition rate, and delay between the laser trigger and MS ion accumulation.
Over the past ten years, transformative developments in the laser technology,[4] data acquisition, motor-control software (RastirX[5]), and imaging processing software (MSiReader[6][7]) have promoted IR-MALDESI as a powerful tool for the direct analysis and MSI of diverse biological, forensic, and pharmaceutical samples.
Principles of operation
In a typical MALDESI MS experiment, a thin layer of ice is deposited on the sample as the energy-absorbing medium. A mid-IR laser[4] that excites the O-H stretching mode of water is used to desorb the neutral materials from the biological samples. The plume of desorbed compounds interacts with an orthogonal electrospray plume where they partition into charged electrospray droplets and are ionized via a process similar to ESI. The ions are subsequently introduced into a mass spectrometer.[2] The ESI-like ionization mechanism has been experimentally demonstrated and studied in depth,[8][9][10] showing equivalent softness as ESI and enabling the detection of intact protein complexes.
The formation of ice matrix starts with purging the enclosure to below 12% relative humidity with the dry nitrogen gas source prior to cooling the Peltier stage down to −10 °C. Afterwards, the cooled sample is exposed to ambient relative humidity, causing the rapid formation of an ice layer on the sample. The ice thickness is maintained throughout the entire experiment by keeping the relative humidity of the enclosure at 10%±2% via the nitrogen purged enclosure.[2] Previously, ice was used as a matrix in IR-MALDI experiments;[11] however, the ion yields for such experiments have been very low. The electrospray post-ionization employed in IR-MALDESI helps alleviate issues associated with low ionization yield. An exogenously deposited ice matrix has been shown to improve the ion abundance by a factor of approximately 15.[12]
MALDESI is a complex interactive system, making it challenging to generate the optimum geometry in just one single experiment. The experimental settings involved in the communication between the sampling stage, mass spectrometer and laser desorption, such as stage height, ESI-Spot distance and laser repetition rate, have undergone progressive optimization by several design of experiments (DOE).[13][9][2] In addition to the source geometry optimization, the electrospray solvent composition has an effect on the MALDESI signals (i.e. influencing molecular coverage and ion abundance). In a study to improve the detection of tissue-specific lipids, the electrospray parameters have been tailored for positive and negative ionization polarities. Under the optimal parameters, lipid abundances increased by 3-fold with 15% greater coverage in positive polarity, while in negative mode lipid abundance achieved 1.5-fold increase with 10% more coverage.[14]
While the first generation of MALDESI system was coupled to Fourier transform (FT) ion cyclotron resonance mass spectrometer,[3] the current source is interfaced with a Thermo Exploris 240 or a Q Exactive Plus mass spectrometer equipped with an Orbitrap mass analyzer. These mass analyzers offer equally accurate mass measurements but generally shorten the data acquisition duration. MALDESI source can also be coupled to drift tube ion mobility spectrometer-mass spectrometer (IMS-MS) for high-throughput screening.[15] These integrations provide more reliable raw data for MSI as well as direct analysis of various biological specimens because long analysis time could cause physiological changes to the samples under interrogation.
MSiReader
MSiReader is a Matlab application, providing visualization and analyses of high-resolution accurate mass data collected via MALDESI. MSiReader was developed at North Carolina State University and was first released in 2013.[6] It has now become one of the most important free, open-source software options for MSI data,[7] and is compatible with most common MSI data formats (e.g., mzXML, imzML, img, ASCII). There are many essential functions available in MSiReader, namely heat map generation with high mass measurement accuracy,[7] peak normalization,[16] absolute and relative quantification,[7][17] and polarity switching.[18] Advanced features like principal component analysis (MSiPCA), MSiCorrelation,[19] and 3D visualization[20][21] have also been added to MSiReader. To date, MSiReader is used by over 1250 researchers and cited in more than 325 publications since 2013. It is released under the BSD 3 open-source license and can be downloaded freely from the public MSiReader website www.msireader.com. A standalone version that does not require a Matlab license is also available.
RastirX
The Rastir software, also built on a Matlab platform, was developed to enable users to visually define a rectangular region-of-interest (ROI) surrounding a tissue section for MALDESI experiments. Motion control commands and laser and instrument synchronization parameters are automatically generated by Rastir to move the sample stage under the laser beam to acquire the MS data voxel-by-voxel. The latest software version, denoted RastirX, allows users to draw arbitrary ROI on a live video image of a sample with the computer mouse. The ROI can be further modified scan-by-scan, which leads to a significantly shorter acquisition time and less contamination from off-tissue compounds than using a rectangular ROI. This arbitrary ROI tool has been applied in several projects.[5][22]
Applications
Since its debut in 2006, MALDESI has experienced great advancements in laser technology (from UV lasers to various mid-IR wavelength lasers),[1][4][23] ESI settings[9][13][14] and associated software.[5][6][7][8][9] All the progress has made IR-MALDESI a competitive imaging and direct analysis tool to investigate a wide variety of biological samples. By associating m/z with positions where they are acquired, unique ion maps of analytes can be generated, which provides valuable information for spatial distributions of lipids, peptides, metabolomics, and other small biomolecules from mammalian samples[1][12][21][23][14][24] to plant tissues.[25][26][27]
Biological
Proteins and peptides
The ESI-like process enabled MALDESI to detect multiply charged peptides and proteins, which could not be realized by MALDI because MALDI generates primarily singly- and doubly-charged ions.[1]
Lipids and metabolomics
IR-MALDESI has been successfully applied for visualizing lipid distributions on the whole-body level, namely neonatal mouse[24] and zebrafish.[28] By employing oversampling method[29] where the stage moving distance is smaller than the laser beam diameter, IR-MALDESI MSI at the cellular level was achieved at a spot-to-spot distance of ~10 micrometer.[30] Moreover, the lipidomic detectability of IR-MALDESI at the single-cell level has been demonstrated with isolated HeLa cell.[31]
In addition to soft tissues (e.g., hen ovaries[32] and rat liver tissues[10]), IR-MALDESI is capable to detect metabolites from unmodified healthy and stroke-affected mouse bones embedded in plaster of Paris, resulting in putatively annotated 826 and 669 tissue-specific species.[22]
IR-MALDESI is an invaluable tool to study metabolism of plant and vegetable samples, as these samples are naturally rich in water. An extensive metabolomic analysis of cherry tomatoes by IR-MALDESI was recently reported.[25] In the analysis of Arabidopsis seedlings, the amounts of auxin-related compounds, was relatively quantified using stable-isotope-labelled (SIL) indole-3-acetic acid (IAA). Moreover, in this study, agarose was used as an appropriate substrate for first time used in IR-MALDESI.[27]
Neurotransmitters
IR-MALDESI has shown the ability to measure selected neurotransmitters in rat brains exposed to flame retardant tetrabromobisphenol A without any chemical derivatization.[33] The follow-up work that focused on neurotransmitters and their pathway-related metabolites in rat placenta sections reported 49 putatively identified neurotransmitters and metabolites.[34]
Glycans
Glycans are structurally complex molecules that pose many analytical challenges. The direct analysis of N-linked glycans from bovine fetuin by IR-MALDESI in both positive and negative ionization modes was presented, which indicates that IR-MALDESI can be an alternative technique for N-linked glycans profiling.[35]
Three dimensional MSI
Although most traditional 3D MSI build on reconstruction of 2D images of analytes from serial sections, the serial-section-based 3D imaging is time-consuming and can lose significant biological information. IR-MALDESI is an ablation-based technique, which is able to measure and generate 3D heat maps of biomolecules from pharmaceutical tablets[20] and nude mouse skin[21] by sequentially imaging a sample with consecutive ablation events.
Pharmaceutical
In a proof-of-concept study, HIV antiretroviral drugs in incubated cervical tissues were imaged and quantified using IR-MALDESI, and the results were evaluated by a validated LC-MS/MS method.[12] So far, the pharmaceutical application of IR-MALDESI has extended from tissue sections[12][24][36] to single hair strands.[17]
Forensic
Textile fibers are an important form of trace evidence for criminal investigation. One of the characteristics of fibers that can be used to distinguish the victim from the suspect is color generated by a fiber dye along with its associated impurities.[37] A direct analysis of the dye from the fabric was successfully performed using IR-MALDESI without prior separation by chromatography.[38][39]
Related techniques
There are other hybrid ionization methods which combine resonant or non-resonant laser desorption with post electrospray ionization. One example is electrospray laser desorption ionization (ELDI), which uses an ultraviolet laser to form ions by irradiating the sample directly, then interacting with the electrospray plume without using any matrices.[40] The infrared version of ELDI has been referred to as laser ablation electrospray ionization (LAESI). IR-MALDESI differs from these two methods as an exogenous ice matrix is used to enhance the desorption of the sample as well as the geometric parameters of the source. Another technique, desorption atmospheric pressure photoionization (DAPPI), uses a jet of heated solvent vapor to desorb analytes from the sample surface via a nebulizer microchip. The desorbed molecules are then ionized by photons emitted by a UV lamp and introduced to mass spectrometers.[41]
References
- ↑ 1.0 1.1 1.2 1.3 Sampson, Jason S.; Hawkridge, Adam M.; Muddiman, David C. (December 2006). "Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) fourier transform ion cyclotron resonance mass spectrometry". Journal of the American Society for Mass Spectrometry 17 (12): 1712–1716. doi:10.1016/j.jasms.2006.08.003. ISSN 1044-0305. PMID 16952462.
- ↑ 2.0 2.1 2.2 2.3 Robichaud, Guillaume; Barry, Jeremy A.; Muddiman, David C. (2014-01-03). "IR-MALDESI Mass Spectrometry Imaging of Biological Tissue Sections Using Ice as a Matrix". Journal of the American Society for Mass Spectrometry 25 (3): 319–328. doi:10.1007/s13361-013-0787-6. ISSN 1044-0305. PMID 24385399. Bibcode: 2014JASMS..25..319R.
- ↑ 3.0 3.1 Robichaud, Guillaume; Barry, Jeremy A.; Garrard, Kenneth P.; Muddiman, David C. (2012-12-04). "Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FT-ICR Mass Spectrometer". Journal of the American Society for Mass Spectrometry 24 (1): 92–100. doi:10.1007/s13361-012-0505-9. ISSN 1044-0305. PMID 23208743.
- ↑ 4.0 4.1 4.2 Ekelöf, Måns; Manni, Jeffrey; Nazari, Milad; Bokhart, Mark; Muddiman, David C. (2018-02-17). "Characterization of a novel miniaturized burst-mode infrared laser system for IR-MALDESI mass spectrometry imaging". Analytical and Bioanalytical Chemistry 410 (9): 2395–2402. doi:10.1007/s00216-018-0918-9. ISSN 1618-2642. PMID 29455285.
- ↑ 5.0 5.1 5.2 Garrard, Kenneth P.; Ekelöf, Måns; Khodjaniyazova, Sitora; Bagley, M. Caleb; Muddiman, David C. (2020-12-02). "A Versatile Platform for Mass Spectrometry Imaging of Arbitrary Spatial Patterns". Journal of the American Society for Mass Spectrometry 31 (12): 2547–2552. doi:10.1021/jasms.0c00128. ISSN 1044-0305. PMID 32539373. PMC 8761386. https://doi.org/10.1021/jasms.0c00128.
- ↑ 6.0 6.1 6.2 Robichaud, Guillaume; Garrard, Kenneth P.; Barry, Jeremy A.; Muddiman, David C. (2013-03-28). "MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform". Journal of the American Society for Mass Spectrometry 24 (5): 718–721. doi:10.1007/s13361-013-0607-z. ISSN 1044-0305. PMID 23536269. Bibcode: 2013JASMS..24..718R.
- ↑ 7.0 7.1 7.2 7.3 7.4 Bokhart, Mark T.; Nazari, Milad; Garrard, Kenneth P.; Muddiman, David C. (2017-09-20). "MSiReader v1.0: Evolving Open-Source Mass Spectrometry Imaging Software for Targeted and Untargeted Analyses". Journal of the American Society for Mass Spectrometry 29 (1): 8–16. doi:10.1007/s13361-017-1809-6. ISSN 1044-0305. PMID 28932998.
- ↑ 8.0 8.1 Dixon, R. Brent; Muddiman, David C. (2010). "Study of the ionization mechanism in hybrid laser based desorption techniques". The Analyst 135 (5): 880–2. doi:10.1039/b926422a. ISSN 0003-2654. PMID 20419234. Bibcode: 2010Ana...135..880D. http://dx.doi.org/10.1039/b926422a.
- ↑ 9.0 9.1 9.2 9.3 Rosen, Elias P.; Bokhart, Mark T.; Ghashghaei, H. Troy; Muddiman, David C. (2015-04-04). "Influence of Desorption Conditions on Analyte Sensitivity and Internal Energy in Discrete Tissue or Whole Body Imaging by IR-MALDESI". Journal of the American Society for Mass Spectrometry 26 (6): 899–910. doi:10.1007/s13361-015-1114-1. ISSN 1044-0305. PMID 25840812. Bibcode: 2015JASMS..26..899R.
- ↑ 10.0 10.1 Tu, Anqi; Muddiman, David C. (2019-09-09). "Internal Energy Deposition in Infrared Matrix-Assisted Laser Desorption Electrospray Ionization With and Without the Use of Ice as a Matrix". Journal of the American Society for Mass Spectrometry 30 (11): 2380–2391. doi:10.1007/s13361-019-02323-2. ISSN 1044-0305. PMID 31502226. Bibcode: 2019JASMS..30.2380T.
- ↑ Nelson, R.; Rainbow, M.; Lohr, D.; Williams, P (1989-12-22). "Volatilization of high molecular weight DNA by pulsed laser ablation of frozen aqueous solutions". Science 246 (4937): 1585–1587. doi:10.1126/science.2595370. ISSN 0036-8075. PMID 2595370. Bibcode: 1989Sci...246.1585N. http://dx.doi.org/10.1126/science.2595370.
- ↑ 12.0 12.1 12.2 12.3 Barry, Jeremy A.; Robichaud, Guillaume; Bokhart, Mark T.; Thompson, Corbin; Sykes, Craig; Kashuba, Angela D. M.; Muddiman, David C. (2014-04-18). "Mapping Antiretroviral Drugs in Tissue by IR-MALDESI MSI Coupled to the Q Exactive and Comparison with LC-MS/MS SRM Assay". Journal of the American Society for Mass Spectrometry 25 (12): 2038–2047. doi:10.1007/s13361-014-0884-1. ISSN 1044-0305. PMID 24744212. Bibcode: 2014JASMS..25.2038B.
- ↑ 13.0 13.1 Barry, Jeremy A.; Muddiman, David C. (2011-11-08). "Global optimization of the infrared matrix-assisted laser desorption electrospray ionization (IR MALDESI) source for mass spectrometry using statistical design of experiments". Rapid Communications in Mass Spectrometry 25 (23): 3527–3536. doi:10.1002/rcm.5262. ISSN 0951-4198. PMID 22095501. Bibcode: 2011RCMS...25.3527B.
- ↑ 14.0 14.1 14.2 Bagley, M. Caleb; Ekelöf, Måns; Muddiman, David C. (2020-02-05). "Determination of Optimal Electrospray Parameters for Lipidomics in Infrared-Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry Imaging". Journal of the American Society for Mass Spectrometry 31 (2): 319–325. doi:10.1021/jasms.9b00063. ISSN 1044-0305. PMID 32031399. https://doi.org/10.1021/jasms.9b00063.
- ↑ Ekelöf, Måns; Dodds, James; Khodjaniyazova, Sitora; Garrard, Kenneth P.; Baker, Erin S.; Muddiman, David C. (2020-03-04). "Coupling IR-MALDESI with Drift Tube Ion Mobility-Mass Spectrometry for High-Throughput Screening and Imaging Applications". Journal of the American Society for Mass Spectrometry 31 (3): 642–650. doi:10.1021/jasms.9b00081. ISSN 1044-0305. PMID 31971795.
- ↑ Tu, Anqi; Muddiman, David C. (2019-06-25). "Systematic evaluation of repeatability of IR-MALDESI-MS and normalization strategies for correcting the analytical variation and improving image quality". Analytical and Bioanalytical Chemistry 411 (22): 5729–5743. doi:10.1007/s00216-019-01953-5. ISSN 1618-2642. PMID 31240357.
- ↑ 17.0 17.1 Rosen, Elias P.; Thompson, Corbin G.; Bokhart, Mark T.; Prince, Heather M. A.; Sykes, Craig; Muddiman, David C.; Kashuba, Angela D. M. (2016-01-19). "Analysis of Antiretrovirals in Single Hair Strands for Evaluation of Drug Adherence with Infrared-Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry Imaging". Analytical Chemistry 88 (2): 1336–1344. doi:10.1021/acs.analchem.5b03794. ISSN 0003-2700. PMID 26688545.
- ↑ Nazari, Milad; Muddiman, David C. (2016-01-04). "Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI)" (in en). Analyst 141 (2): 595–605. doi:10.1039/C5AN01513H. ISSN 1364-5528. PMID 26402586. Bibcode: 2016Ana...141..595N.
- ↑ Ekelöf, Måns; Garrard, Kenneth P.; Judd, Rika; Rosen, Elias P.; Xie, De-Yu; Kashuba, Angela D. M.; Muddiman, David C. (2018-12-01). "Evaluation of Digital Image Recognition Methods for Mass Spectrometry Imaging Data Analysis". Journal of the American Society for Mass Spectrometry 29 (12): 2467–2470. doi:10.1007/s13361-018-2073-0. ISSN 1044-0305. PMID 30324263. Bibcode: 2018JASMS..29.2467E.
- ↑ 20.0 20.1 Bai, Hongxia; Khodjaniyazova, Sitora; Garrard, Kenneth P.; Muddiman, David C. (2020-02-05). "Three-Dimensional Imaging with Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry". Journal of the American Society for Mass Spectrometry 31 (2): 292–297. doi:10.1021/jasms.9b00066. ISSN 1044-0305. PMID 32031410. PMC 8284694. https://doi.org/10.1021/jasms.9b00066.
- ↑ 21.0 21.1 21.2 Bai, Hongxia; Linder, Keith E.; Muddiman, David C. (2021-01-02). "Three-dimensional (3D) imaging of lipids in skin tissues with infrared matrix-assisted laser desorption electrospray ionization (MALDESI) mass spectrometry" (in en). Analytical and Bioanalytical Chemistry 413 (10): 2793–2801. doi:10.1007/s00216-020-03105-6. ISSN 1618-2650. PMID 33388847.
- ↑ 22.0 22.1 Khodjaniyazova, Sitora; Hanne, Nicholas J.; Cole, Jacqueline H.; Muddiman, David C. (2019-11-28). "Mass spectrometry imaging (MSI) of fresh bones using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI)" (in en). Analytical Methods 11 (46): 5929–5938. doi:10.1039/C9AY01886G. ISSN 1759-9679. PMID 33815571.
- ↑ 23.0 23.1 Sampson, Jason S.; Murray, Kermit K.; Muddiman, David C. (2009-04-01). "Intact and top-down characterization of biomolecules and direct analysis using infrared matrix-assisted laser desorption electrospray ionization coupled to FT-ICR mass spectrometry". Journal of the American Society for Mass Spectrometry 20 (4): 667–673. doi:10.1016/j.jasms.2008.12.003. ISSN 1044-0305. PMID 19185512.
- ↑ 24.0 24.1 24.2 Bokhart, Mark T.; Rosen, Elias; Thompson, Corbin; Sykes, Craig; Kashuba, Angela D. M.; Muddiman, David C. (2015-03-01). "Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization" (in en). Analytical and Bioanalytical Chemistry 407 (8): 2073–2084. doi:10.1007/s00216-014-8220-y. ISSN 1618-2650. PMID 25318460.
- ↑ 25.0 25.1 Bagley, M. Caleb; Pace, Crystal L.; Ekelöf, Måns; Muddiman, David C. (2020). "Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) mass spectrometry imaging analysis of endogenous metabolites in cherry tomatoes". The Analyst 145 (16): 5516–5523. doi:10.1039/d0an00818d. ISSN 0003-2654. PMID 32602477. Bibcode: 2020Ana...145.5516B.
- ↑ Kalmar, Jaclyn Gowen; Oh, Yeonyee; Dean, Ralph A.; Muddiman, David C. (2019-11-23). "Investigating host-pathogen meta-metabolic interactions of Magnaporthe oryzae infected barley using infrared matrix-assisted laser desorption electrospray ionization mass spectrometry". Analytical and Bioanalytical Chemistry 412 (1): 139–147. doi:10.1007/s00216-019-02216-z. ISSN 1618-2642. PMID 31760448. http://dx.doi.org/10.1007/s00216-019-02216-z.
- ↑ 27.0 27.1 Bagley, M. Caleb; Stepanova, Anna N.; Ekelöf, Måns; Alonso, Jose M.; Muddiman, David C. (2020). "Development of a relative quantification method for infrared matrix-assisted laser desorption electrospray ionization mass spectrometry imaging of Arabidopsis seedlings" (in en). Rapid Communications in Mass Spectrometry 34 (6): e8616. doi:10.1002/rcm.8616. ISSN 1097-0231. PMID 31658400.
- ↑ Stutts, Whitney L.; Knuth, Megan M.; Ekelöf, Måns; Mahapatra, Debabrata; Kullman, Seth W.; Muddiman, David C. (2020-04-01). "Methods for Cryosectioning and Mass Spectrometry Imaging of Whole-Body Zebrafish". Journal of the American Society for Mass Spectrometry 31 (4): 768–772. doi:10.1021/jasms.9b00097. ISSN 1044-0305. PMID 32129621. PMC 9375048. https://doi.org/10.1021/jasms.9b00097.
- ↑ Jurchen, John C.; Rubakhin, Stanislav S.; Sweedler, Jonathan V. (October 2005). "MALDI-MS imaging of features smaller than the size of the laser beam". Journal of the American Society for Mass Spectrometry 16 (10): 1654–1659. doi:10.1016/j.jasms.2005.06.006. ISSN 1044-0305. PMID 16095912.
- ↑ Nazari, Milad; Muddiman, David C. (2015-03-01). "Cellular-level mass spectrometry imaging using infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) by oversampling" (in en). Analytical and Bioanalytical Chemistry 407 (8): 2265–2271. doi:10.1007/s00216-014-8376-5. ISSN 1618-2650. PMID 25486925.
- ↑ Xi, Ying; Tu, Anqi; Muddiman, David C. (2020-11-01). "Lipidomic profiling of single mammalian cells by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI)" (in en). Analytical and Bioanalytical Chemistry 412 (29): 8211–8222. doi:10.1007/s00216-020-02961-6. ISSN 1618-2650. PMID 32989513.
- ↑ Lakeh, Mahsa Akbari; Tu, Anqi; Muddiman, David C.; Abdollahi, Hamid (2019). "Discriminating normal regions within cancerous hen ovarian tissue using multivariate hyperspectral image analysis" (in en). Rapid Communications in Mass Spectrometry 33 (4): 381–391. doi:10.1002/rcm.8362. ISSN 1097-0231. PMID 30468547. Bibcode: 2019RCMS...33..381A. https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/abs/10.1002/rcm.8362.
- ↑ Bagley, M. Caleb; Ekelöf, Måns; Rock, Kylie; Patisaul, Heather; Muddiman, David C. (2018-12-01). "IR-MALDESI mass spectrometry imaging of underivatized neurotransmitters in brain tissue of rats exposed to tetrabromobisphenol A" (in en). Analytical and Bioanalytical Chemistry 410 (30): 7979–7986. doi:10.1007/s00216-018-1420-0. ISSN 1618-2650. PMID 30317443.
- ↑ Pace, Crystal L.; Horman, Brian; Patisaul, Heather; Muddiman, David C. (2020-06-01). "Analysis of neurotransmitters in rat placenta exposed to flame retardants using IR-MALDESI mass spectrometry imaging" (in en). Analytical and Bioanalytical Chemistry 412 (15): 3745–3752. doi:10.1007/s00216-020-02626-4. ISSN 1618-2650. PMID 32300844.
- ↑ Pace, Crystal L.; Muddiman, David C. (2020-08-05). "Direct Analysis of Native N-Linked Glycans by IR-MALDESI". Journal of the American Society for Mass Spectrometry 31 (8): 1759–1762. doi:10.1021/jasms.0c00176. ISSN 1044-0305. PMID 32603137. PMC 8285077. https://doi.org/10.1021/jasms.0c00176.
- ↑ Thompson, Corbin G.; Bokhart, Mark T.; Sykes, Craig; Adamson, Lourdes; Fedoriw, Yuri; Luciw, Paul A.; Muddiman, David C.; Kashuba, Angela D. M. et al. (2015-05-01). "Mass Spectrometry Imaging Reveals Heterogeneous Efavirenz Distribution within Putative HIV Reservoirs" (in en). Antimicrobial Agents and Chemotherapy 59 (5): 2944–2948. doi:10.1128/AAC.04952-14. ISSN 0066-4804. PMID 25733502.
- ↑ Goodpaster, John V.; Liszewski, Elisa A. (2009-08-01). "Forensic analysis of dyed textile fibers" (in en). Analytical and Bioanalytical Chemistry 394 (8): 2009–2018. doi:10.1007/s00216-009-2885-7. ISSN 1618-2650. PMID 19543886. https://doi.org/10.1007/s00216-009-2885-7.
- ↑ Cochran, Kristin H.; Barry, Jeremy A.; Muddiman, David C.; Hinks, David (2013-01-15). "Direct Analysis of Textile Fabrics and Dyes Using Infrared Matrix-Assisted Laser Desorption Electrospray Ionization Mass Spectrometry". Analytical Chemistry 85 (2): 831–836. doi:10.1021/ac302519n. ISSN 0003-2700. PMID 23237031.
- ↑ Cochran, Kristin H.; Barry, Jeremy A.; Robichaud, Guillaume; Muddiman, David C. (2015-01-01). "Analysis of trace fibers by IR-MALDESI imaging coupled with high resolving power MS" (in en). Analytical and Bioanalytical Chemistry 407 (3): 813–820. doi:10.1007/s00216-014-8042-y. ISSN 1618-2650. PMID 25081013.
- ↑ Shiea, Jentaie; Huang, Min-Zon; HSu, Hsiu-Jung; Lee, Chi-Yang; Yuan, Cheng-Hui; Beech, Iwona; Sunner, Jan (2005). "Electrospray-assisted laser desorption/ionization mass spectrometry for direct ambient analysis of solids" (in en). Rapid Communications in Mass Spectrometry 19 (24): 3701–3704. doi:10.1002/rcm.2243. ISSN 1097-0231. PMID 16299699. Bibcode: 2005RCMS...19.3701S. https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/abs/10.1002/rcm.2243.
- ↑ Haapala, Markus; Pól, Jaroslav; Saarela, Ville; Arvola, Ville; Kotiaho, Tapio; Ketola, Raimo A.; Franssila, Sami; Kauppila, Tiina J. et al. (2007-10-01). "Desorption Atmospheric Pressure Photoionization". Analytical Chemistry 79 (20): 7867–7872. doi:10.1021/ac071152g. ISSN 0003-2700. PMID 17803282. https://doi.org/10.1021/ac071152g.
 |