Chemistry:Aurintricarboxylic acid
| |
| |
| Names | |
|---|---|
| Preferred IUPAC name
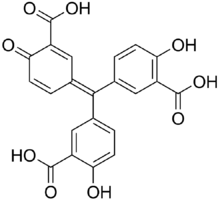
3,3′-[(3-Carboxy-4-oxocyclohexa-2,5-dien-1-ylidene)methylene]bis(6-hydroxybenzoic acid) | |
| Other names
5,5′-[(3-Carboxy-4-oxocyclohexa-2,5-dien-1-ylidene)methylene]bis(2-hydroxybenzoic acid)
Aluminon free acid | |
| Identifiers | |
3D model (JSmol)
|
|
| Abbreviations | ATA |
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
PubChem CID
|
|
| UNII | |
| |
| |
| Properties | |
| C22H14O9 | |
| Molar mass | 422.345 g·mol−1 |
| Appearance | Dark red to brown powder |
| Melting point | 300 °C (572 °F; 573 K) |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Aurintricarboxylic acid (ATA) is a chemical compound that readily polymerizes in aqueous solution, forming a stable free radical that inhibits protein-nucleic acid interactions. It is a potent inhibitor of ribonuclease and topoisomerase II by preventing the binding of the nucleic acid to the enzyme. It stimulates tyrosine phosphorylation processes including the Jak2/STAT5 pathway in NB2 lymphoma cells, ErbB4 in neuroblastoma cells, and MAP kinases, Shc proteins, phosphatidylinositide 3-kinase and phospholipase Cγ in PC12 cells. It also inhibits apoptosis. It prevents down-regulation of Ca2+-impermeable GluR2 receptors and inhibits calpain, a Ca2+-activated protease that is activated during apoptosis.[1]
It is used to inhibit protein biosynthesis in its initial stages. Nominally, it is used in biological experiments as a protein inhibitor, and as an ammonium salt (known as aluminon) it is used as a reagent to estimate the aluminium in water, biological tissue, and foods.[2]
It was found that ATA is a strong inhibitor of topoisomerases and other nucleases. It might be useful for increasing efficiency of RNA isolation.[3]
It has been discovered that using aurintricarboxylic acid against influenza-A post-infection has a strong protective effect by inhibiting the virus' ability to reproduce. In cultured canine kidney cells, it was found to reduce viral reproduction and infection when applied post-infection, but not when used as a 'vaccine'.[4] It has also been shown to block the binding of the HIV coat molecule gp120 to the CD4 co-receptor on T cells through which it invades.[citation needed]
Aurintricarboxylic acid and its ammonium salt shows antiviral activity in vitro against coronaviruses such as SARS, MERS and SARS-CoV-2, and while it is unlikely to have suitable properties to be developed as a medicine in its own right, it has proved useful in scientific research into novel antiviral drugs to combat these diseases.[5][6]
Preparation
Aurintricarboxylic acid can be prepared by the condensation of formaldehyde with salicylic acid in the presence of nitrite-containing sulfuric acid.[7]
References
- ↑ 1.0 1.1 "Aurintricarboxylic acid". Sigma-Aldrich. http://www.sigmaaldrich.com/catalog/product/sigma/a1895?lang=en.
- ↑ "Aurintricarboxylic Acid". Reference.MD. http://www.reference.md/files/D001/mD001312.html.
- ↑ "Characterization and use of the potent ribonuclease inhibitor aurintricarboxylic acid for the isolation of RNA from animal tissues". The Biochemical Journal 263 (1): 73–80. October 1989. doi:10.1042/bj2630073. PMID 2481441.
- ↑ "Aurintricarboxylic acid is a potent inhibitor of influenza A and B virus neuraminidases". PLOS ONE 4 (12): e8350. December 2009. doi:10.1371/journal.pone.0008350. PMID 20020057. Bibcode: 2009PLoSO...4.8350H.
- ↑ "Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases". ACS Central Science 6 (3): 315–331. 2020. doi:10.1021/acscentsci.0c00272. PMID 32226821.
- ↑ "Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV". ChemBioChem 21 (5): 730–738. March 2020. doi:10.1002/cbic.202000047. PMID 32022370.
- ↑ Vogel, Arthur I.. Practical Organic Chemistry Including Qualitative Organic Analysis.
 |

