Biology:Oxidosqualene cyclase
Oxidosqualene cyclases (OSC) are enzymes involved in cyclization reactions of 2,3-oxidosqualene to form sterols or triterpenes.[1]
There are two major groups of sterol-producing OSC enzymes:
- Cycloartenol synthase (CAS), found in all plants, which produces primarily cycloartenol
- Lanosterol synthase (LAS), found in all animals and fungi, and occasionally in plants, which produces primarily lanosterol
Sterols and triterpenes are extremely diverse classes of natural products, particularly in plants, which often contain numerous OSC enzymes with different substrate and product specificities;[1] common examples include lupeol synthase and beta-amyrin synthase.[2] OSC enzymes' catalytic mechanism is similar to the prokaryotic squalene-hopene cyclase.[3]
Directed evolution and protein design have been used to identify small numbers of point mutations that alter the product specificities of OSC enzymes, most notably in altering a cycloartenol synthase to produce predominantly lanosterol.[4]
Structure
Oxidosqualene cyclase is a monomeric enzyme. Its active site consists of a depression between two barrel domains.[5] The active site is mostly made up of acidic amino acids in the majority of organisms.[6] The residues in the active site make it energetically favorable for oxidosqualene to take on a more folded conformation, which closely resembles its product.[5] This crucially sets the substrate up for the series of reactions that form the rings. Oxidosqualene is located in the cell’s microsome membranes where it can easily harvest its hydrophobic substrate and turn out its hydrophobic product.[7]
Function
Oxidosqualene cyclase is a key enzyme in the cholesterol biosynthesis pathway. It catalyzes the formation of lanosterol, which is then converted through many steps into cholesterol. The body uses cholesterol for temperature regulation. It is also a precursor for testosterone in males and oestradiol in females.[8]
Regulation
The enzyme’s genetic expression is regulated by sterol regulatory element binding protein (SREBP-2), a molecule which also regulates the expression of other enzymes in the cholesterol biosynthesis pathway.[9] The SREBP-2 transcription factor increases enzymatic activity upstream of OSC, such as that of HMG-CoA reductase and squalene synthase, to increase flux through sterol synthesis process.[10]
Mechanism
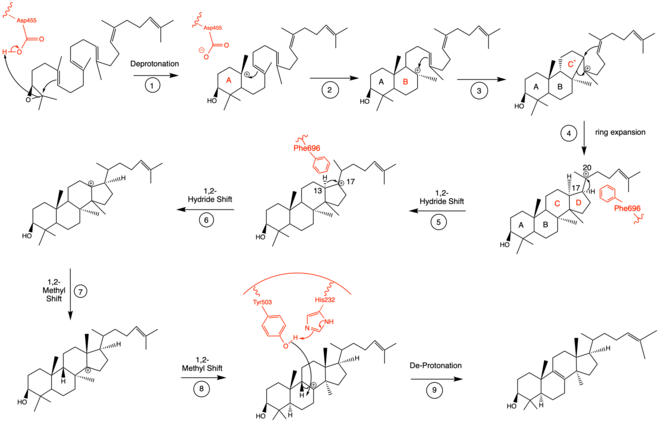
Mechanistically, the enzyme oxidosqualene:lanosterol cyclase catalyzes the formation of four rings along the long chain of the substrate (oxidosqualene), producing lanosterol. This cyclization is one of the most complex known enzyme functions[11] and is highly selective.[12] The reaction can be characterized by two main mechanistic series: an epoxide ring opening followed by a series of ring closures and a series of 1,2-hydride and 1,2-methyl shifts. These mechanistic steps are catalyzed by a collection of amino acids in the active site.[13]
Ring closure
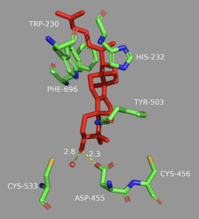
In the enzyme’s active site, a histidine residue activates an aspartic acid residue (which has been identified as Asp455),[13] which protonates the substrate’s epoxide, setting off a series of carbon-carbon bond formations that form rings.[7][14] More specifically, this ring closure occurs ring by ring (labeled A-D in the mechanism below). It is widely accepted that the epoxide protonation is concurrent with the A ring closure.[15] The cation is then transferred from C10 to C8 to form the B ring.[15] It is the C ring formation that puzzles scholars. However, recent studies have come to a conclusion that the ring is first closed via a Markovnikov addition, forming a 5-carbon intermediate.[15] Following this, the D ring is formed, pushing out the C ring to its 6-carbon final state.[15] To note, QM/MM studies have shown that the protonation of this epoxide ring is the rate limiting step of the entire mechanism,[13] indicating it may be the most regulated.
Hydride and methyl shifts
The intermediate formed after the four ring closures undergoes two 1,2-hydride shifts and two 1,2-methyl shifts to be in position for final de-protonation.[16] Molecular dynamics simulations have proved critical to understanding the conformational changes that oxidosqualene-lanosterol intermediates undergo.[16] Researchers have identified Phe696 as a critical amino acid for controlled and specific hydride shifting in this mechanism (steps 5-6), as it speeds up the shift by lowering the energy barrier of the respective transition states.[16]
Final de-protonation
Finally, the enzyme de-protonates to yield lanosterol, which has a hydroxyl group instead of an epoxide. This hydroxyl group can be seen in the image above. The literature has shown that two amino acids in OSC are critical for this de-protonation to occur correctly and uniformly. Tyrosine at the 503rd position works in tandem with histidine at the 203rd position to de-protonate the hydrogen at the carbon adjacent to the carbocation in intermediate 8.[13] This is accomplished as shown in the proposed mechanism below. The crystal structure of the OSC-lanosterol complex confirms that Tyr503 and His232 are in optimal positions for this final de-protonation step.[13]
Disease relevance
High blood cholesterol, also called hypercholesterolemia, significantly increases the risk of stroke, heart attack, and peripheral artery disease. If untreated, it can also lead to plaque accumulation in blood vessels, which is known as atherosclerosis.[17] For this reason, the sterol biosynthetic pathway has long been a target for the drug development industry. Statins, which inhibit HMG-CoA reductase (an enzyme that catalyzes an earlier step in the cholesterol biosynthesis pathway) are commonly prescribed to treat high cholesterol. Research is being done for other compounds which block different steps in the biosynthesis of cholesterol, including the reaction performed by oxidosqualene cyclase which cyclizes squalene to form lanosterol.[7]
Oxidosqualene cyclase, which is downstream of squalene in the pathway, is an attractive target for inhibition. Many inhibitors have been proposed, among them steroid analogs, phenol-based compounds, benzamide and carboxamide derivatives, and nitrogen-containing heterocyclic compounds. The most effective inhibitors have a hydrogen-bond acceptor at a specific distance away from a hydrophobic region.[6]
Inhibitors of oxidosqualene cyclase have shown promise as antimicrobial agents as well, because they’ve been shown to kill off trypanosoma cruzi.[18][19] Trypanosoma cruzi is a parasite transmitted to people by insects, mostly in Latin America. The parasite causes a disease called Chagas disease, in which acute infections around an insect bite can lead to more serious complications, such as decreased heart, esophagus, colon, and even brain function.[20] Another parasite, Leishmania donovani, is the causative agent for leishmaniasis, a similar tropical disease that is spread by sand flies and disproportionately affects rural areas.[21] GSK2920487A, a cyclic and aromatic small molecule, has been shown to significantly inhibit oxidosqualene cyclase and decrease the survival of the L. donovani.[22] While there is concern that parasites such as T. cruzi and L. donovani are able to scavenge the sterol compounds that oxidosqualene cyclase catalyzes the formation of, the potency of chemical inhibition against this vital enzyme demonstrate the potential of OSC as a therapeutic target.
Evolution
Stork, et al. compared the protein sequences of C. albicans oxidosqualene cyclase with the analogous enzyme (squalene cyclase) in two different bacteria and found conserved regions in the former.[23] Rabelo et al. found a conserved active site across seven organisms.[6] The specific function and regulation of OSC seems to deviate even among eukaryotes. For example, OSC in both mammals and yeast are accompanied by a ketoreductase protective enzyme. However, this enzyme is necessary for OSC function in yeast only, suggesting the divergent evolution of the steroid biosynthesis pathway in mammals.[24] It is believed that animal and fungal oxidosqualene cyclases likely evolved from their prokaryotic counterparts.[23]
While the biosynthetic steps and catalytic amino acids of human OSC have been characterized, less is known about OSC specificity in other kingdoms of life. For example, the oomycete Saprolegnia parasitica, a fish pathogen, possesses a unique amino acid triad present in only some plant LAS enzymes, though the triad is responsible for determining CAS of LAS specificity in this class of organisms.[25]
References
- ↑ 1.0 1.1 "Triterpene biosynthesis in plants". Annual Review of Plant Biology 65 (1): 225–257. 29 April 2014. doi:10.1146/annurev-arplant-050312-120229. PMID 24498976.
- ↑ "Plant lanosterol synthase: divergence of the sterol and triterpene biosynthetic pathways in eukaryotes". Plant & Cell Physiology 47 (5): 673–677. May 2006. doi:10.1093/pcp/pcj032. PMID 16531457.
- ↑ "Structure and function of a squalene cyclase". Science 277 (5333): 1811–1815. September 1997. doi:10.1126/science.277.5333.1811. PMID 9295270.
- ↑ "Enzyme redesign: two mutations cooperate to convert cycloartenol synthase into an accurate lanosterol synthase". Journal of the American Chemical Society 127 (41): 14132–14133. October 2005. doi:10.1021/ja053791j. PMID 16218577.
- ↑ 5.0 5.1 "Lord of the rings--the mechanism for oxidosqualene:lanosterol cyclase becomes crystal clear". Trends in Pharmacological Sciences 26 (7): 335–340. July 2005. doi:10.1016/j.tips.2005.05.004. PMID 15951028.
- ↑ 6.0 6.1 6.2 "Design strategies of oxidosqualene cyclase inhibitors: Targeting the sterol biosynthetic pathway". The Journal of Steroid Biochemistry and Molecular Biology 171: 305–317. July 2017. doi:10.1016/j.jsbmb.2017.05.002. PMID 28479228.
- ↑ 7.0 7.1 7.2 "Oxidosqualene Cyclase". PDB101: Molecule of the Month. RCSB Protein Data Bank. December 2007. http://pdb101.rcsb.org/motm/96.
- ↑ Biochemistry (7th ed.). New York: W.H. Freeman and Company. 2012. ISBN 978-1-4292-7635-1.
- ↑ "Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes". Proceedings of the National Academy of Sciences of the United States of America 100 (21): 12027–12032. October 2003. doi:10.1073/pnas.1534923100. PMID 14512514. Bibcode: 2003PNAS..10012027H.
- ↑ "Cholesterol synthesis-related enzyme oxidosqualene cyclase is required to maintain self-renewal in primary erythroid progenitors". Cell Proliferation 44 (5): 441–452. October 2011. doi:10.1111/j.1365-2184.2011.00771.x. PMID 21951287.
- ↑ "Metabolism of structural lipids". Lipid Biochemistry: An Introduction. Springer US. 1991. pp. 295–337. doi:10.1007/978-1-4615-3862-2_7. ISBN 9781461538622.
- ↑ Bloch, Konrad. “The Biological Synthesis of Cholesterol.” Nobel Lecture, December 11, 1964.
- ↑ 13.0 13.1 13.2 13.3 13.4 "Catalytic mechanism and product specificity of oxidosqualene-lanosterol cyclase: a QM/MM study". The Journal of Physical Chemistry B 116 (47): 13857–13862. November 2012. doi:10.1021/jp3091396. PMID 23130825.
- ↑ "Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase". Nature 432 (7013): 118–122. November 2004. doi:10.1038/nature02993. PMID 15525992. Bibcode: 2004Natur.432..118T.
- ↑ 15.0 15.1 15.2 15.3 "Concerted Cyclization of Lanosterol C-Ring and D-Ring Under Human Oxidosqualene Cyclase Catalysis: An ab Initio QM/MM MD Study". Journal of Chemical Theory and Computation 10 (3): 1109–1120. March 2014. doi:10.1021/ct400949b. PMID 26580186.
- ↑ 16.0 16.1 16.2 "Biosynthetic Mechanism of Lanosterol: A Completed Story" (in en). ACS Catalysis 10 (3): 2157–2168. 2020-02-07. doi:10.1021/acscatal.9b05221. ISSN 2155-5435.
- ↑ "High Blood Cholesterol". National Heart, Lung, and Blood Institute (NHLBI). https://www.nhlbi.nih.gov/health-topics/high-blood-cholesterol.
- ↑ "Oxidosqualene cyclase inhibitors as antimicrobial agents". Journal of Medicinal Chemistry 46 (20): 4240–4243. September 2003. doi:10.1021/jm034126t. PMID 13678402.
- ↑ "Potent anti-Trypanosoma cruzi activities of oxidosqualene cyclase inhibitors". Antimicrobial Agents and Chemotherapy 45 (4): 1210–1215. April 2001. doi:10.1128/AAC.45.4.1210-1215.2001. PMID 11257036.
- ↑ "Chagas Disease". U.S. Centers for Disease Control and Prevention. 2017-05-02. https://www.cdc.gov/parasites/chagas/disease.html.
- ↑ "CDC - Leishmaniasis - General Information - Frequently Asked Questions (FAQs)" (in en-us). U.S. Centers for Disease Control and Prevention. 2020-05-19. https://www.cdc.gov/parasites/leishmaniasis/gen_info/faqs.html.
- ↑ "Multiple unbiased approaches identify oxidosqualene cyclase as the molecular target of a promising anti-leishmanial" (in English). Cell Chemical Biology 28 (5): 711–721.e8. May 2021. doi:10.1016/j.chembiol.2021.02.008. PMID 33691122.
- ↑ 23.0 23.1 "The Stereochemistry of Polyene Cyclization". Journal of the American Chemical Society 77 (19): 5068–5077. October 1955. doi:10.1021/ja01624a038. ISSN 0002-7863.
- ↑ "Divergent interactions involving the oxidosqualene cyclase and the steroid-3-ketoreductase in the sterol biosynthetic pathway of mammals and yeasts". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1801 (11): 1232–1237. November 2010. doi:10.1016/j.bbalip.2010.07.006. PMID 20659585.
- ↑ "The Oxidosqualene Cyclase from the Oomycete Saprolegnia parasitica Synthesizes Lanosterol as a Single Product". Frontiers in Microbiology 7: 1802. 2016. doi:10.3389/fmicb.2016.01802. PMID 27881978.
External links
- Overview of all the structural information available in the PDB for UniProt: P48449 (Lanosterol synthase) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: P04035 (HMG-CoA reductase) at the PDBe-KB.
 |