Biology:Phage P22 tailspike protein
| Bifunctional tail protein | |
|---|---|
 | |
| Identifiers | |
| Organism | |
| Symbol | 9 |
| Entrez | 1262799 |
| PDB | 2XC1 (ECOD) |
| RefSeq (mRNA) | NC_002371.2 |
| RefSeq (Prot) | NP_059644.1 |
| Other data | |
| EC number | 3.2.1.- |
| Chromosome | Genome: 0.02 - 0.02 Mb |
The tailspike protein (P22TSP) of Enterobacteria phage P22 mediates the recognition and adhesion between the bacteriophage and the surface of Salmonella enterica cells. It is anchored within the viral coat and recognizes the O-antigen portion of the lipopolysaccharide (LPS) on the outer-membrane of Gram-negative bacteria. It possesses endoglycanase activity, serving to shorten the length of the O-antigen during infection.[2]
History
The initial interest in tailspike proteins was in the study of the effect the mutations on protein folding.[3] Some mutations affect the folding efficiency of the protein but have no effect on the final native structure. Other mutations have been identified that lead to a temperature sensitive phenotype.[4] Reconstitution experiments have demonstrated that the in vitro folding process closely mirrors the in vivo folding pathway.[3] It has been further been demonstrated that folding yields in vitro decrease strongly with increasing temperature.[5]
Function
O-antigen binding
P22TSP recognizes the O-antigen polysaccharide of LPS serotypes A, B, or D1. The serotypes correspond to species S. Typhimurium, S. Enteritidis, and S. Paratyphi A.[6] These carbohydrates share the same main chain trisaccharide repeating unit alpha-D-mannose-(1—4)-alpha-L-rhamnose-(1—3)-alpha-D-galactose-(1—2), but each have a different 2,6-dideoxyhexose substituent at C-3 of the mannose.[7]
In vivo, P22TSP binds as a homotrimer and one phage particle can carry up to 6 tailspikes. P22TSP can bind multivalently, leading to an essentially irreversible attachment.[7] It was shown that a minimum of two repeating units or an octasaccharide is required for binding.[2] The TSP is also capable of binding longer fragments with similar affinity.
Endoglycosidase activity
P22TSP has endorhamnosidase activity and cleaves the glycosidic bond of the rhamnose group, producing an octasaccharide product. Two aspartic acids and one glutamic acid in the active site have been strongly linked to enzymatic activity.[2] Different biological functions for this cleavage have been proposed. Cleavage could facilitate access to the membrane.[8] or allow the phage to find the optimal position for infection.[9]
Role in DNA injection
It has been demonstrated that cleavage of the O-antigen is necessary for DNA ejection by the phage. It has been proposed that P22TSP binding positions the phage to inject its DNA.[10]
Structure
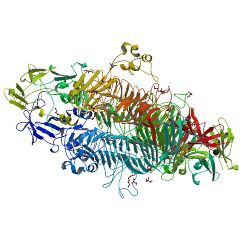
P22TSP is a homotrimeric structural protein consisting of 666 amino acids. It is noncovalently bound to the neck of the viral capsid. It has been crystallized in space group P213 and has one monomer in the asymmetric unit. The secondary structure of P22TSP is dominated by a parallel Beta helix comprising 13 complete turns.[11] This structure is further characterized as a beta-solenoid domain.
P22TSP is compOsed of two domains, each with distinct function. An N-terminal domains serves to bind to the phage particle and a C-terminal domains that interacts with the Salmonella surface. These two domains are connected by a flexible linker.[2]
The binding site of P22TSP is located in the central part of the Beta helix. A deep cleft is formed by a 60-residue insertion on one side along with three smaller 5-25 residue insertions on the other.[12]
Homologous proteins
Several functional homologues of P22TSP has been identified belonging to the bacteriophages HK620 and Sf6. Both of these tailspike proteins also contain right-handed parallel beta-helices and share similar O-antigen binding and cleavage to P22TSP. These proteins share 70% sequence identity in their N-terminal domains, but no sequence similarities have been found in the C-terminal domains.[13]
Translational applications
Carbohydrate binding scaffolds
P22TSP has also been studied due to its high kinetic stability. As it exists and functions in the extracellular environment, it must endure harsh conditions such as highly variable temperatures or high concentrations of protein degrading enzymes.[14] The kinetic stability of P22TSP derives from its compact beta-solenoid architecture. It was shown that like other viral fibrous proteins, P22 tailspike protein possesses a high stability against denaturation. This makes P22TSP a promising candidate for use as thermostable scaffold capable of being tailor-made to recognize heteropolymers.[14]
Therapy against Salmonella infection
Tailspike proteins have also shown potential for more translational applications such as fighting bacterial infections. A study has demonstrated that orally administered P22TSP markedly reduced Salmonella colonization in a group of chickens. They suggest that the endorhamnosidase activity of the free tailspike molecule serves to modify O-antigen, compromising the LPS structure and thereby preventing the binding of a phage P22-attached tailspike protein. The authors suggest that this has the potential to be a novel therapy meant to fight bacterial infections.[15]
References
- ↑ 2XC1, 2VFM, 2VKY, 2VFP, 2VFQ, 2VFO, 2VFN, 2VNL, 3TH0, 1TYU, 1TYV, 1TYX, 1TYW, 1TSP, 1CLW, 1QRB, 1QRC, 1QQ1, 1QA1, 1QA2, 1QA3, 1LKT
- ↑ 2.0 2.1 2.2 2.3 "Carbohydrate binding of Salmonella phage P22 tailspike protein and its role during host cell infection". Biochemical Society Transactions 38 (5): 1386–1389. Oct 2010. doi:10.1042/BST0381386. PMID 20863318.
- ↑ 3.0 3.1 "Mechanism of phage P22 tailspike protein folding mutations". Protein Science 2 (11): 1869–1881. Nov 1993. doi:10.1002/pro.5560021109. PMID 8268798.
- ↑ "Amino acid substitutions influencing intracellular protein folding pathways". FEBS Letters 307 (1): 20–25. Jul 1992. doi:10.1016/0014-5793(92)80894-M. PMID 1639189.
- ↑ "Folding and assembly of phage P22 tailspike endorhamnosidase lacking the N-terminal, head-binding domain". European Journal of Biochemistry 215 (3): 653–661. Aug 1993. doi:10.1111/j.1432-1033.1993.tb18076.x. PMID 8354271.
- ↑ "An essential serotype recognition pocket on phage P22 tailspike protein forces Salmonella enterica serovar Paratyphi A O-antigen fragments to bind as nonsolution conformers". Glycobiology 23 (4): 486–494. Apr 2013. doi:10.1093/glycob/cws224. PMID 23292517.
- ↑ 7.0 7.1 "Enthalpic barriers to the hydrophobic binding of oligosaccharides to phage P22 tailspike protein". Biochemistry 40 (17): 5144–5150. May 2001. doi:10.1021/bi0020426. PMID 11318636.
- ↑ "Bacterial surface carbohydrates and bacteriophage adsorption". Surface Carbohydrates of the Procaryotic Cell: 289–356. 1977.
- ↑ "Effects of receptor destruction by Salmonella bacteriophages epsilon 15 and c341". Virology 105 (2): 328–337. Sep 1980. doi:10.1016/0042-6822(80)90034-3. PMID 7423851.
- ↑ "Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro". The Journal of Biological Chemistry 285 (47): 36768–36775. Nov 2010. doi:10.1074/jbc.M110.169003. PMID 20817910.
- ↑ "Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer". Science 265 (5170): 383–386. Jul 1994. doi:10.1126/science.8023158. PMID 8023158. Bibcode: 1994Sci...265..383S.
- ↑ "Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors". Proceedings of the National Academy of Sciences of the United States of America 93 (20): 10584–10588. Oct 1996. doi:10.1073/pnas.93.20.10584. PMID 8855221. Bibcode: 1996PNAS...9310584S.
- ↑ "Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related". Molecular Microbiology 69 (2): 303–316. Jul 2008. doi:10.1111/j.1365-2958.2008.06311.x. PMID 18547389.
- ↑ 14.0 14.1 "Phage tailspike proteins with beta-solenoid fold as thermostable carbohydrate binding materials". Macromolecular Bioscience 9 (2): 169–173. Feb 2009. doi:10.1002/mabi.200800278. PMID 19148901.
- ↑ "Orally administered P22 phage tailspike protein reduces salmonella colonization in chickens: prospects of a novel therapy against bacterial infections". PLOS ONE 5 (11): e13904. 2010. doi:10.1371/journal.pone.0013904. PMID 21124920. Bibcode: 2010PLoSO...513904W.
 |

