Biology:DNA-binding protein from starved cells
| Dps (DNA-binding proteins from starved cells) | |
|---|---|
 | |
| Identifiers | |
| Symbol | DPS |
| InterPro | IPR002177 |
| CDD | cd01043 |
DNA-binding proteins from starved cells (DPS) are bacterial proteins that belong to the ferritin superfamily and are characterized by strong similarities but also distinctive differences with respect to "canonical" ferritins.
DPS proteins are part of a complex bacterial defence system that protects DNA against oxidative damage and are distributed widely in the bacterial kingdom.
Description
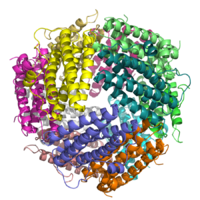
DPS are highly symmetrical dodecameric proteins of 20 kDa characterized from a shell-like structure of 2:3 tetrahedral symmetry assembled from identical subunits with an external diameter of ~ 9 nm and a central cavity of ~ 4.5 nm in diameter.[2][3][4] Dps proteins belong to the ferritin superfamily and the DNA protection is afforded by means of a double mechanism:
The first was discovered in Escherichia coli Dps in 1992 [5] and has given the name to the protein family; during stationary phase, Dps binds the chromosome non-specifically, forming a highly ordered and stable Dps-DNA co-crystal within which chromosomal DNA is condensed and protected from diverse damages.[6] The lysine-rich N-terminus is required for self-aggregation as well as for Dps-driven DNA condensation.[7]
The second mode of protection is due to the ability of Dps proteins to bind and oxidize Fe(II) at the characteristic, highly conserved intersubunit ferroxidase center.[8][9]
The dinuclear ferroxidase centers are located at the interfaces between subunits related by 2-fold symmetry axes.[10] Fe(II) is sequestered and stored in the form of Fe(III) oxyhydroxide mineral, which can be released after reduction. In the mineral iron core up to 500 Fe(III) can be deposited. One hydrogen peroxide oxidizes two Fe2+ ions, which prevents hydroxyl radical production by the Fenton reaction (reaction I):
2 Fe2+ + H2O2 + 2 H+ = 2 Fe3+ + 2 H2O
Dps also protects the cell from UV and gamma ray irradiation, iron and copper toxicity, thermal stress and acid and base shocks.[1] Also shows a weak catalase activity.
DNA condensation
Dps dodecamers can condense DNA in vitro through a cooperative binding mechanism. Deletion of portions of the N-terminus[7] or mutation of key lysine residues in the N-terminus[11] can impair or eliminate the condensation activity of Dps. Single molecule studies have shown that Dps-DNA complexes can get trapped in long-lived metastable states that exhibit hysteresis.[12] Because of this, the extent of DNA condensation by Dps can depend not only on the current buffer conditions but also on the conditions in the past. A modified Ising model can be used to explain this binding behavior.
Expression
In Escherichia coli Dps protein is Induced by rpoS and IHF in the early stationary phase. Dps is also Induced by oxyR in response to oxidative stress during exponential phase. ClpXP probably directly regulate proteolysis of dps during exponential phase. ClpAP seems to play an indirect role in maintaining ongoing dps synthesis during stationary phase
Applications
For nanoparticle synthesis
Cavities formed by Dps and ferritin proteins have been successfully used as the reaction chamber for the fabrication of metal nanoparticles (NPs).[13][14][15][16] Protein shells served as a template to restrain particle growth and as a coating to prevent coagulation/aggregation between NPs. Using various sizes of protein shells, various sizes of NPs can be easily synthesized for chemical, physical and bio-medical applications.
For enzyme encapsulation
Nature utilizes protein-based architectures to house enzymes within its interior cavity, for example: encapsulin and carboxysomes. Taking inspiration from nature, hollow interior cavity of Dps and ferritin cages have also been used to encapsulate enzymes.[17] Cytochrome C, a hemoprotein with peroxidase-like activity when encapsulated inside Dps cage showed better catalytic activity over broad pH range compared to free enzyme in bulk solution. This behavior was attributed to high local concentration of enzyme inside Dps and unique microenvironment provided by Dps interior cavity.[18]
For targeted drug delivery
Delivery of cargo at intended target site remains major concern for targeted drug delivery owing to presence of biological barriers and enhanced permeability and retention (EPR) effects. Furthermore, formation of protein corona around injected nanoparticles is also a topic of interest within the targeted delivery field. Researchers tried to overcome these concerns by using natural bio-distribution of protein cage nanoparticles for cargo delivery. For example, DNA binding protein from nutrient starved cells (Dps) cage was shown to cross glomerular filtration barrier and target renal proximal tubules.[19]
See also
References
- ↑ 1.0 1.1 "The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site". Nature Structural Biology 7 (1): 38–43. January 2000. doi:10.1038/71236. PMID 10625425.
- ↑ "The crystal structure of Dps, a ferritin homolog that binds and protects DNA". Nature Structural Biology 5 (4): 294–303. April 1998. doi:10.1038/nsb0498-294. PMID 9546221.
- ↑ "The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding". Biochimica et Biophysica Acta (BBA) - General Subjects 1800 (8): 798–805. August 2010. doi:10.1016/j.bbagen.2010.01.013. PMID 20138126.
- ↑ "Role of Dps (DNA-binding proteins from starved cells) aggregation on DNA". Frontiers in Bioscience 15 (1): 122–31. January 2010. doi:10.2741/3610. PMID 20036810.
- ↑ "A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli". Genes & Development 6 (12B): 2646–54. December 1992. doi:10.1101/gad.6.12b.2646. PMID 1340475.
- ↑ "DNA protection by stress-induced biocrystallization". Nature 400 (6739): 83–5. July 1999. doi:10.1038/21918. PMID 10403254. Bibcode: 1999Natur.400...83W.
- ↑ 7.0 7.1 "DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus". Nucleic Acids Research 32 (19): 5935–44. 2004. doi:10.1093/nar/gkh915. PMID 15534364.
- ↑ "Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli". The Journal of Biological Chemistry 277 (31): 27689–96. August 2002. doi:10.1074/jbc.M202094200. PMID 12016214.
- ↑ "The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: x-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties". The Journal of Biological Chemistry 278 (22): 20319–26. May 2003. doi:10.1074/jbc.M302114200. PMID 12660233.
- ↑ "Dps protects cells against multiple stresses during stationary phase". Journal of Bacteriology 186 (13): 4192–8. July 2004. doi:10.1128/JB.186.13.4192-4198.2004. PMID 15205421.
- ↑ "The DNA-Binding Protein from Starved Cells (Dps) Utilizes Dual Functions To Defend Cells against Multiple Stresses". Journal of Bacteriology 197 (19): 3206–15. October 2015. doi:10.1128/JB.00475-15. PMID 26216848.
- ↑ "Hysteresis in DNA compaction by Dps is described by an Ising model". Proceedings of the National Academy of Sciences of the United States of America 113 (18): 4982–7. May 2016. doi:10.1073/pnas.1521241113. PMID 27091987. Bibcode: 2016PNAS..113.4982V.
- ↑ "Protein Cage Constrained Synthesis of Ferrimagnetic Iron Oxide Nanoparticles". Advanced Materials 14 (21): 1562–1565. 2002. doi:10.1002/1521-4095(20021104)14:21<1562::AID-ADMA1562>3.0.CO;2-D.
- ↑ "Constrained synthesis of cobalt oxide nanomaterials in the 12-subunit protein cage from Listeria innocua". Inorganic Chemistry 42 (20): 6300–5. October 2003. doi:10.1021/ic0343657. PMID 14514305.
- ↑ "Synthesis of iron oxide nanoparticles in Listeria innocua Dps (DNA-binding protein from starved cells): a study with the wild-type protein and a catalytic centre mutant". Chemistry: A European Journal 16 (2): 709–17. January 2010. doi:10.1002/chem.200901138. PMID 19859920.
- ↑ "Suzuki-Miyaura cross-coupling catalyzed by protein-stabilized palladium nanoparticles under aerobic conditions in water: application to a one-pot chemoenzymatic enantioselective synthesis of chiral biaryl alcohols". Green Chemistry 11 (12): 1929. 2009. doi:10.1039/b915184b.
- ↑ "Enzyme Encapsulation by a Ferritin Cage". Angewandte Chemie 56 (47): 14933–14936. November 2017. doi:10.1002/anie.201708530. PMID 28902449.
- ↑ "Cytochrome C with peroxidase-like activity encapsulated inside the small DPS protein nanocage". Journal of Materials Chemistry B 9: 3168–3179. March 2021. doi:10.1039/d1tb00234a. PMID 33885621.
- ↑ "The archaeal Dps nanocage targets kidney proximal tubules via glomerular filtration". Journal of Clinical Investigation 129: 3941–3951. September 2019. doi:10.1172/JCI127511. PMID 31424427.
 |

