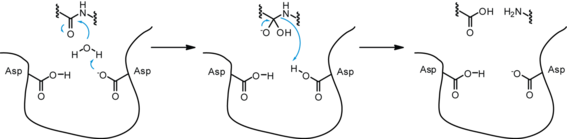Biology:HIV-1 protease
| HIV-1 Protease (Retropepsin) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
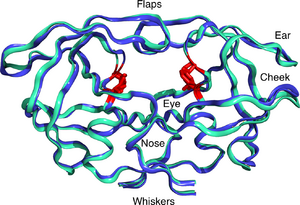
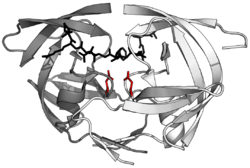 HIV-1 protease dimer in white and grey, with peptide substrate in black and active site aspartate side chains in red. (PDB: 1KJF) | |||||||||
| Identifiers | |||||||||
| EC number | 3.4.23.16 | ||||||||
| CAS number | 144114-21-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
HIV-1 protease or PR is a retroviral aspartyl protease (retropepsin), an enzyme involved with peptide bond hydrolysis in retroviruses, that is essential for the life-cycle of HIV, the retrovirus that causes AIDS.[1][2] HIV-1 PR cleaves newly synthesized polyproteins (namely, Gag and Gag-Pol[3]) at nine cleavage sites to create the mature protein components of an HIV virion, the infectious form of a virus outside of the host cell.[4] Without effective HIV-1 PR, HIV virions remain uninfectious.[5][6]
Structure
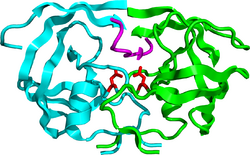
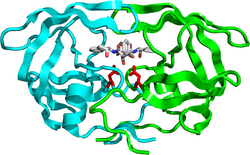
Mature HIV protease exists as a 22 kDa homodimer, with each subunit made up of 99 amino acids.[1] A single active site lies between the identical subunits and has the characteristic Asp-Thr-Gly (Asp25, Thr26 and Gly27) catalytic triad sequence common to aspartic proteases.[8] As HIV-1 PR can only function as a dimer, the mature protease contains two Asp25 amino acids, one from each monomer, that act in conjunction with each other as the catalytic residues.[9] Additionally, HIV protease has two molecular "flaps" which move a distance of up to 7 Å when the enzyme becomes associated with a substrate.[10] This can be visualized with animations of the flaps opening and closing.
Biosynthesis
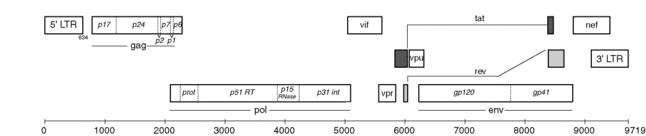
Precursor
The Gag-Pol polyprotein, which contains premature coding proteins, including HIV-1 PR.[9] PR is located between the reverse transcriptase (which is at the C-terminus of PR) and the p6pol (which is at the N-terminus of PR) of the transframe region (TFR).[11]
In order for this precursor to become a functional protein, each monomer must associate with another HIV-1 PR monomer to form a functional catalytic active site by each contributing the Asp25 of their respective catalytic triads.[9]
Synthesis Mechanism
When viral HIV-RNA enters the cell, it is accompanied by a reverse transcriptase, an integrase, and a mature HIV-1 PR. The reverse transcriptase converts viral RNA into DNA, facilitating the integrase's role in incorporating viral genetic information with the host cell DNA.[2] The viral DNA can either remain dormant in the nucleus or be transcribed into mRNA and translated by the host cell into the Gag-Pol polyprotein, which would then be cleaved into individual functional proteins (including a newly synthesized HIV-1 PR) by the mature HIV-1 PR.[9]
The HIV-1 PR precursor catalyzes its own production by facilitating its cleavage from the Gag-Pol polyprotein in a mechanism known as auto-processing. Auto-processing of HIV-1 PR is characterized by two sequential steps: (1) the intramolecular cleavage of the N-terminus at the p6pol-protease cleavage site, which serves to finalize PR processing and increase enzymatic activity with the newly formed PR-reverse transcriptase intermediate, and (2) the intermolecular cleavage of the C-terminus at the protease-reverse transcriptase cleavage site, leading to the assembly of two PR subunits into mature dimers.[12][13] Dimerization of the two subunits allows for fully functional, combined active site, characterized by two Asp25 catalytic residues (one from each monomer), to form.[14]
Function
HIV-1 PR serves a dual purpose. Precursor HIV-1 PR is responsible for catalyzing its own production into mature PR enzymes via PR auto-processing.[15] Mature protease is able to hydrolyze peptide bonds on the Gag-Pol polyproteins at nine specific sites, processing the resulting subunits into mature, fully functional proteins. These cleaved proteins, including reverse transcriptase, integrase, and RNaseH, are encoded by the coding region components necessary for viral replication.[4]
Mechanism
As an aspartic protease, the dimerized HIV-1 PR functions through the aspartyl group complex, in order to perform hydrolysis. Of the two Asp25 residues on the combined catalytic active site of HIV-1 PR, one is deprotonated while the other is protonated, due to pKa differences from the micro-environment.[16]
In a general aspartic protease mechanism, once the substrate is properly bound to the active site of the enzyme, the deprotonated Asp25 catalytic amino acid undergoes base catalysis, rendering the incoming water molecule a better nucleophile by deprotonating it. The resulting hydroxyl ion attacks the carbonyl carbon of the peptide bond, forming an intermediate with a transient oxyanion, which is stabilized by the initially protonated Asp25. The oxyanion re-forms a double bond, leading to the cleavage of the peptide bond between the two amino acids, while the initially deprotonated Asp25 undergoes acid catalysis to donate its proton to the amino group, making the amino group a better leaving group for complete peptide bond cleavage and returning to its original deprotonated state.[2][17]
While HIV-1 PR shares many of the same characteristics as a non-viral aspartic protease, some evidence has shown that HIV-1 PR catalyzes hydrolysis in a concerted manner; in other words, the nucleophilic water molecule and the protonated Asp25 simultaneously attack the scissile peptide bond during catalysis.[17][18]
As a drug target
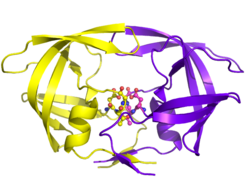
With its integral role in HIV replication, HIV protease has been a prime target for drug therapy. HIV protease inhibitors work by specifically binding to the active site by mimicking the tetrahedral intermediate of its substrate and essentially becoming “stuck,” disabling the enzyme. After assembly and budding, viral particles lacking active protease cannot mature into infectious virions. Several protease inhibitors have been licensed for HIV therapy.[19]
There are ten HIV-1 PR inhibitors that are currently approved by the Food and Drug Administration: indinavir, saquinavir, ritonavir, nelfinavir, lopinavir, amprenavir, fosamprenevir, atazanavir, tipranavir, and darunavir. Many of the inhibitors have different molecular components and thus different mechanistic actions, such as blocking the active site. Their functional roles also extend to influencing circulation concentrations of other inhibitor drugs (ritonavir) and being used only for certain circumstances in which the virus exhibits tolerance of other inhibitors (tipranavir).[4][20]
Evolution and resistance
Due to the high mutation rates of retroviruses, especially due to mutationally sensitive regions (notably the region containing the catalytic triad sequence), and considering that changes to a few amino acids within HIV protease can render it much less visible to an inhibitor, the active site of this enzyme can change rapidly when under the selective pressure of replication-inhibiting drugs.[21][22]
Two types of mutations are generally associated with increasing drug resistance: "major" mutations and "secondary" mutations. Major mutations involve a mutation on the active site of HIV-1 PR, preventing the selective inhibitors from binding it. Secondary mutations refer to molecular changes on the periphery of the enzyme due to prolonged exposure of similar chemicals, potentially affecting inhibitor specificity for HIV-1 PR.[3]
One approach to minimizing the development of drug-resistance in HIV is to administer a combination of drugs which inhibit several key aspects of the HIV replication cycle simultaneously, rather than one drug at a time. Other drug therapy targets include reverse transcriptase, virus attachment, membrane fusion, cDNA integration and virion assembly.[23][24]
See also
- Management of HIV/AIDS
- Discovery and development of HIV-protease inhibitors
External links
- The MEROPS online database for peptidases and their inhibitors: A02.001
- Proteopedia HIV-1_protease - the HIV-1 protease structure in interactive 3D.
- Proteopedia Flaps_Morph_for_HIV_Protease - Animation of the flaps opening and closing based on X-ray crystal structures.
- HIV-1+Protease at the US National Library of Medicine Medical Subject Headings (MeSH)
References
- ↑ 1.0 1.1 "The structure and function of the aspartic proteinases". Annual Review of Biophysics and Biophysical Chemistry 19 (1): 189–215. 1990. doi:10.1146/annurev.bb.19.060190.001201. PMID 2194475. https://zenodo.org/record/1234933.
- ↑ 2.0 2.1 2.2 "HIV-1 protease: mechanism and drug discovery". Organic & Biomolecular Chemistry 1 (1): 5–14. January 2003. doi:10.1039/b208248a. PMID 12929379.
- ↑ 3.0 3.1 "The role of select subtype polymorphisms on HIV-1 protease conformational sampling and dynamics". The Journal of Biological Chemistry 289 (24): 17203–14. June 2014. doi:10.1074/jbc.M114.571836. PMID 24742668.
- ↑ 4.0 4.1 4.2 "HIV protease inhibitors: a review of molecular selectivity and toxicity" (in en). HIV/AIDS: Research and Palliative Care 7: 95–104. April 2015. doi:10.2147/hiv.s79956. PMID 25897264.
- ↑ "Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides". Proceedings of the National Academy of Sciences of the United States of America 86 (3): 807–11. February 1989. doi:10.1073/pnas.86.3.807. PMID 2644644. Bibcode: 1989PNAS...86..807K.
- ↑ "Active human immunodeficiency virus protease is required for viral infectivity". Proceedings of the National Academy of Sciences of the United States of America 85 (13): 4686–90. July 1988. doi:10.1073/pnas.85.13.4686. PMID 3290901. Bibcode: 1988PNAS...85.4686K.
- ↑ "HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs". Protein Science 13 (4): 1108–23. April 2004. doi:10.1110/ps.03468904. PMID 15044738. PMC 2280056. https://ai.stanford.edu/~serafim/CS374_2005/Papers/MolecularDynamics_DrugResistance.pdf.
- ↑ "Folding regulates autoprocessing of HIV-1 protease precursor". The Journal of Biological Chemistry 280 (12): 11369–78. March 2005. doi:10.1074/jbc.M412603200. PMID 15632156.
- ↑ 9.0 9.1 9.2 9.3 "Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism". Journal of Virology 78 (16): 8477–85. August 2004. doi:10.1128/JVI.78.16.8477-8485.2004. PMID 15280456.
- ↑ "Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution". Science 246 (4934): 1149–52. December 1989. doi:10.1126/science.2686029. PMID 2686029.
- ↑ "Autoprocessing of HIV-1 protease is tightly coupled to protein folding" (in En). Nature Structural Biology 6 (9): 868–75. September 1999. doi:10.1038/12327. PMID 10467100.
- ↑ "Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease from an analog of the Gag-Pol polyprotein". Proceedings of the National Academy of Sciences of the United States of America 91 (17): 7970–4. August 1994. doi:10.1073/pnas.91.17.7970. PMID 8058744. Bibcode: 1994PNAS...91.7970L.
- ↑ "A transient precursor of the HIV-1 protease. Isolation, characterization, and kinetics of maturation". The Journal of Biological Chemistry 271 (8): 4477–81. February 1996. doi:10.1074/jbc.271.8.4477. PMID 8626801.
- ↑ "HIV-1 protease function and structure studies with the simplicial neighborhood analysis of protein packing method". Proteins 73 (3): 742–53. November 2008. doi:10.1002/prot.22094. PMID 18498108.
- ↑ "Understanding HIV-1 protease autoprocessing for novel therapeutic development". Future Medicinal Chemistry 5 (11): 1215–29. July 2013. doi:10.4155/fmc.13.89. PMID 23859204.
- ↑ "Ionization states of the catalytic residues in HIV-1 protease". Nature Structural Biology 3 (11): 946–50. November 1996. doi:10.1038/nsb1196-946. PMID 8901873.
- ↑ 17.0 17.1 "A combined quantum/classical molecular dynamics study of the catalytic mechanism of HIV protease". Journal of Molecular Biology 261 (3): 454–69. August 1996. doi:10.1006/jmbi.1996.0476. PMID 8780786.
- ↑ "Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor". Biochemistry 30 (6): 1600–9. February 1991. doi:10.1021/bi00220a023. PMID 1993177.
- ↑ Rang and Dale's pharmacology (6th ed.). Philadelphia, Pa., U.S.A.: Churchill Livingstone/Elsevier. 2007. ISBN 9780808923541.
- ↑ "Influence of drug transport proteins on the pharmacokinetics and drug interactions of HIV protease inhibitors". Journal of Pharmaceutical Sciences 100 (9): 3636–54. September 2011. doi:10.1002/jps.22655. PMID 21698598.
- ↑ "Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors". Antimicrobial Agents and Chemotherapy 47 (2): 759–69. February 2003. doi:10.1128/AAC.47.2.759-769.2003. PMID 12543689.
- ↑ "Complete mutagenesis of the HIV-1 protease" (in En). Nature 340 (6232): 397–400. August 1989. doi:10.1038/340397a0. PMID 2666861. Bibcode: 1989Natur.340..397L.
- ↑ "New targets for inhibitors of HIV-1 replication". Nature Reviews. Molecular Cell Biology 1 (1): 40–9. October 2000. doi:10.1038/35036060. PMID 11413488.
- ↑ "The design of drugs for HIV and HCV". Nature Reviews. Drug Discovery 6 (12): 1001–18. December 2007. doi:10.1038/nrd2424. PMID 18049474.
 |