Biology:Acetyltransferase
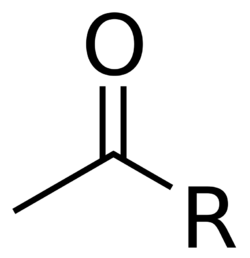
Acetyltransferase (or transacetylase) is a type of transferase enzyme that transfers an acetyl group, through a process called acetylation. Acetylation serves as a modification that can profoundly transform the functionality of a protein by modifying various properties like hydrophobicity, solubility, and surface attributes.[1] These alterations have the potential to influence the protein's conformation and its interactions with substrates, cofactors, and other macromolecules.[1] The image to the right shows the basic structure of an acetyl group, where R is a variable indicates the remainder of the molecule to which the acetyl group is attached.
| Acetyltransferases | Substrate | Gene | Chromosome Location | Gene Group | Abbreviation |
| Histone Acetyltransferase | Lysine residues on histones[1] | HAT1[2] | 2q31.1[2] | Lysine acetyltransferases[2] | HAT |
| Choline Acetyltransferase | Choline[3] | CHAT[4] | 10q11.23[4] | NA | ChAT[3] |
| Serotonin N-Acetyltransferase | Serotonin | AANAT[5] | 17q25.1[5] | GCN5 Related N-Acetyltransferases[5] | AANAT[5] |
| NatA Acetyltransferase | N-terminus of various proteins as they emerge from the ribosome | NAA15[6] | 4q31.1[6] | Armadillo like helical domain containing
N-alpha-acetyltransferase subunits[6] |
NatA[6] |
| NatB Acetyltransferase | Peptides starting with Met-Asp/Glu/Asn/Gln[7] | NAA25[8] | 12q24.13[8] | N-alpha-acetyltransferase subunits
MicroRNA protein coding host genes[8] |
NatB[8] |
Structure
The 3D structure predictions of histone, choline, and serotonin acetyltransferases are shown to the side of this page. The 3D structure of these proteins are essential for interactions between them and their substrates. Alterations to the 3D structures of these enzymes could result in the chemical modifications not being completed.
Additional examples include:
See also
References
- ↑ 1.0 1.1 1.2 "Writers and readers of histone acetylation: structure, mechanism, and inhibition". Cold Spring Harbor Perspectives in Biology 6 (7): a018762. July 2014. doi:10.1101/cshperspect.a018762. PMID 24984779.
- ↑ 2.0 2.1 2.2 "Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase". Current Biology 8 (2): 96–108. January 1998. doi:10.1016/s0960-9822(98)70040-5. PMID 9427644.
- ↑ 3.0 3.1 "Substrate binding and catalytic mechanism of human choline acetyltransferase". Biochemistry 45 (49): 14621–14631. December 2006. doi:10.1021/bi061536l. PMID 17144655.
- ↑ 4.0 4.1 "Human choline acetyltransferase gene maps to region 10q11-q22.2 by in situ hybridization". Genomics 9 (2): 396–398. February 1991. doi:10.1016/0888-7543(91)90273-H. PMID 1840566.
- ↑ 5.0 5.1 5.2 5.3 "The human serotonin N-acetyltransferase (EC 2.3.1.87) gene (AANAT): structure, chromosomal localization, and tissue expression". Genomics 34 (1): 76–84. May 1996. doi:10.1006/geno.1996.0243. PMID 8661026.
- ↑ 6.0 6.1 6.2 6.3 "Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans". Proceedings of the National Academy of Sciences of the United States of America 106 (20): 8157–8162. May 2009. doi:10.1073/pnas.0901931106. PMID 19420222. Bibcode: 2009PNAS..106.8157A.
- ↑ "Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB". Structure 25 (4): 641–649.e3. April 2017. doi:10.1016/j.str.2017.03.003. PMID 28380339.
- ↑ 8.0 8.1 8.2 8.3 "N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB". Proceedings of the National Academy of Sciences of the United States of America 109 (31): 12449–12454. July 2012. doi:10.1073/pnas.1210303109. PMID 22814378. Bibcode: 2012PNAS..10912449V.
External links
- Acetyltransferases at the US National Library of Medicine Medical Subject Headings (MeSH)
 |



