Biology:C16orf71
Uncharacterized protein Chromosome 16 Open Reading Frame 71 is a protein in humans, encoded by the C16orf71 gene.[1] The gene is expressed in epithelial tissue of the respiratory system, adipose tissue, and the testes.[2] Predicted associated biological processes of the gene include regulation of the cell cycle, cell proliferation, apoptosis, and cell differentiation in those tissue types.[3] 1357 bp of the gene are antisense to spliced genes ZNF500 and ANKS3, indicating the possibility of regulated alternate expression.[4]
Gene
Locus
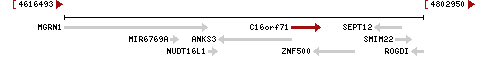
The gene is located on the short arm of chromosome 16 at 16p13.1.[5] Its genomic sequence begins on the plus strand at 4,734,242 bp and ends at 4,749,396 bp.[1]
mRNA
Alternative Splicing
Three different protein encoding transcript variants, or isoforms, have been identified for C16orf71.[7] One non-protein coding transcript variant was identified for the gene.[8]
| Name | Length (bp) | Protein (aa) | Mass (kDa) | Biotype |
|---|---|---|---|---|
| Uncharacterized protein C16orf71 (primary assembly)[7] | 2716 | 520 | 55.7 | Protein coding |
| Uncharacterized protein C16orf71 isoform X2[9] | 2324 | 136 | 14.6 | Protein coding |
| Uncharacterized protein C16orf71 isoform X3[10] | 2435 | 156 | 16.8 | Protein coding |
| Uncharacterized protein C16orf71 isoform X1[11] | 2562 | 537 | 57.5 | Protein coding |
| Uncharacterized protein C16orf71 Transcript-003[8] | 3705 | No protein | – | Retained intron |
Protein
thumb|Evidence of localization at nuclear speckles of the nucleus, indicated by the green spots where in situ hybridization occurred with the antibody.[12]|147x147px
General properties
The primary encoded protein consists of 520 amino acid residues, 11 total exons, and is 15.14 kb long, with a molecular weight of approximately 55.68 kDa.[1] The predicted isoelectric point was reported to be 4.81, indicating it is relatively unstable.[13] The gene was reported to be well expressed, at 1.1 times the average gene level.[4]
Composition
Alanine was the most abundant amino acid, contributing to 11.54% of the molecular weight of the protein.[13] Serine was the second most abundant, contributing 10.19% to the overall molecular weight.[13] The average Alanine frequency in vertebrate proteins is approximately 7.4% and the average Serine frequency is approximately 8.1%.[14]
Domains
C16orf71 has one identified domain of unknown function, DUF4701, that is conserved in all mammals and some species of reptiles and birds.[1] DUF4701 spans from amino acid residue 21 to 520 in the protein.[1]
Post-translational modifications
C16orf71 is predicted to undergo multiple post-translational modifications such as phosphorylation, N-glycosylation, and amidation.
Protein Interactions
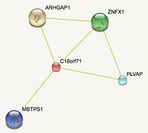
Experimentally proven interactions
Experimentation with C16orf71 has revealed interactions with four other proteins, ARHGAP1, ZNFX1, PLVAP, and MBTPS1.[15] ARHGAP1, ZNFX1, and MBTPS1 are associated with regulation in signaling and metabolism while PLVAP is associated with the formation of small lipid rafts in the plasma membrane of vertebrate endothelial and adipose cells.[3]
Predicted interactions
The majority of the predicted interactions involved with the protein related to regulation of mitotic processes, cellular differentiation, proliferation, metabolism, and signaling.[3] Additional related processes included the formation and differentiation of B cells, T cells, endothelial cells, endoderm, and endocrine glands.[3]
| Interactor[3] | Function[3] |
|---|---|
| CREB1 (cAMP responsive element binding protein 1) | Induction of growth, differentiation, migration, adhesion, and cell survival in epidermal cells
Mediation of growth, differentiation, survival, and migration in early developmental stages Mediation of metabolic functions, tissue repair, and regeneration in mature adult tissue |
| TYK2 (tyrosine kinase 2) | Cellular differentiation, migration, and proliferation in immune cells |
| TNIP2 (TNPAIP3 interacting protein 2) | Negative regulation of apoptosis for endothelial cells |
| OBSL1 (obscurin-like 1) | Mitotic regulation, cytoskeleton and microtubule organization and assembly |
| DUSP3 (dual specificity phosphatase 3) | Negative regulation of multiple enzymatic cascades and signaling pathways
Positive regulation of the mitotic cell cycle |
| FGFRL1 (fibroblast growth factor receptor-like 1) | Fibroblast growth activity |
| GNPAT (glyceronephosphate O-acyltransferase) | Involved in multiple metabolic and biosynthesis processes for cellular lipids, ether lipids,
glycerophospholipids, phosphatidic acid, and phospholipids |
| AURKA (aurora kinase A) | Regulation for G2/M transition, nuclear division, mitotic spindle organization, the centrosome
cycle, cytokinesis, and spindle stabilization |
| NAMPT (nicotinamide phosphoribosyltransferase) | Adipose tissue development, regulation of nicotinamide metabolism, signal transduction,
cell-cell signaling, and vitamin metabolism. |
Subcellular localization
C16orf71 was observed in nuclear speckles of the nucleus through experimental protocols involving fluorescent in situ hybridization with antibodies.[2] Nuclear speckles, also known as interchromatin granule clusters, are enriched in pre-mRNA splicing factors.[16] These highly dynamic structures are located in interchromatin regions of the nucleoplasm in mammalian cells and have been observed to cycle throughout various nuclear regions and active transcription sites.[16]
Structure
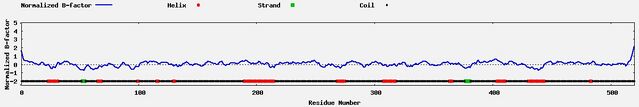
The secondary structure of C16orf71 is predicted to consist primarily of coils, with small regions of alpha helices and two segments of beta sheets throughout the span of the protein.[13][17]
Protein sequences of the gene's mammalian orthologs were analyzed to reveal similar results, while distant reptilian and avian ortholog sequences predicted more regions of beta sheets.[18][19]
Expression

Tissue expression pattern
Human expression for the gene has been observed primarily in respiratory epithelial tissue, specifically the trachea, larynx, nasopharynx, and bronchus.[2] C16orf71 is also moderately expressed in adipose tissue and testes.[2]
DNA microarray experimental data
DNA microarray analysis from various experiments provided information on the expression levels of C16orf71 in unique, varying conditions.
The gene appears to have higher levels of expression in the omental adipose tissue of obese subjects compared to non-obese subjects.[20]


C16orf71 was also observed to have decreased expression when there was a depletion of HIF-1 alpha, HIF-2 beta, or both. HIF, or hypoxia-inducible factors, are responsible for the mediation of hypoxia effects within the body.[23] In addition, HIFs promote clotting and restoration of various epithelial tissues and are vital in the development of mammalian embryos, sperm, and ova.[24]
Data from an experiment also indicated noticeably lower expression of the gene in sperm affected with teratozoospermia, a condition where sperm have abnormal morphology affecting the fertility in males, compared to normal sperm.[22]
C16orf71 was observed to be present in all stages of development, with similar levels of expression throughout.[25]
Toxicogenomics experimental data
Three chemicals, bisphenol A, butyraldehyde, and polychlorinated biphenyls, have been experimentally tested with C16orf71 for evidence of interaction.[26]
Bisphenol A is suspected to cause impairment in male reproduction.[27] An experiment utilizing seminiferous tubule culture was conducted to observe the effects on meiosis and potential germ-line abnormalities.[27] Gene expression analysis revealed decrease expression for C16orf71 when exposed to the chemical.[27]
Butyraldehyde has been observed to affect inflammatory responses in bronchial airway tissue on a genetic level.[28] Microarray analysis was used to determine levels of expression in human alveolar epithelial cells after exposure to the compound.[28] Results indicated decreased expression for C16orf71 when exposed to the chemical.[28]
Polychlorinated biphenyl was used in an experiment to determine its effects on external male genital development.[29] Human fetal corpora cavernosa cells were used as the model tissue.[29] Toxicogenomic analysis indicated the chemical affected all genes involved with genitourinary development and revealed lowered expression levels for C16orf71.[29]
Regulation of expression
1357 bp of the gene are antisense to spliced genes ZNF500 and ANKS3, indicating possibility of regulated alternate expression.[4] A ZNF500 transcription factor binding domain was found on the minus strand within the promoter region of the gene.[30] ZNF500 is predicted to play a role in gene regulation, transcription, and cellular differentiation.[31]
The beginning of the promoter region was predicted to be 117 bp upstream from the 5' UTR of C16orf71 and is 1371 bp long.[30] The region was analyzed for predicted transcription factors and regulatory elements. Predicted transcription factors in the promoter region related to the regulation of the cell cycle, proliferation, apoptosis, and differentiation of sperm and epithelial tissue components.[3]
Predicted transcription factors
| Transcription factor[30] | Associated functions[30] |
|---|---|
| Ascl1 (Mammaliam achaete scute homolog 1) | B-cell differentiation, maturation, and development
Negative regulation of transcription and apoptosis Positive regulation of cell cycle and cellular differentiation Response to hypoxia and epidermal growth factor Regulation of epithelial cell differentiation |
| ZNF500 (Zinc finger with KRAB and SCAN domains 3) | Cartilage development
Negative regulation of gene expression and cellular senescence T-cell and stem cell differentiation Positive regulation of transcription |
| SMAD4 transcription factor involved in TGF-beta signaling | Regulation of apoptosis, T-cell and endothelial cell activation
Endoderm formation and development Negative regulation of cell growth and death Response to hypoxia Thyroid gland development Tissue morphogenesis |
| Cysteine-serine-rich nuclear protein 1 | TGF-beta induced apoptosis
Regulation of early development and differentiation Extracellular matrix formation |
Homology
Paralogs
No human paralogs for the gene were found.[32]
Orthologs
Orthologs have been identified in most mammals for which complete genome data is available.[32] C16orf71 and its domain of unknown function, DUF4701, was present in mammals.[32] The most distant orthologs identified were reptilian.[32][33]
Molecular evolution
The m value, or number of corrected amino acid changes per 100 residues, for the gene C16orf71 was plotted against the divergence of species in millions of years. When compared to the data of hemoglobin, fibrinopeptides, and cytochrome C, it was determined that the gene has the closest progression to fibrinopeptides, suggesting a relatively rapid pace of evolution. M values for C16orf71 were derived from percentage of identity of species mRNA sequences compared to the human sequence using the formula derived from the Molecular Clock Hypothesis.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "C16orf71 Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=C16orf71.
- ↑ 2.0 2.1 2.2 2.3 "Tissue expression of C16orf71 - Summary - The Human Protein Atlas". http://www.proteinatlas.org/ENSG00000166246-C16orf71/tissue.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 "C16orf71 protein (Homo sapiens) - STRING network view". http://string-db.org/cgi/network.pl?taskId=hF14TqV075cr.
- ↑ 4.0 4.1 4.2 Thierry-Mieg, Danielle; Thierry-Mieg, Jean. "AceView: Gene:C16orf71, a comprehensive annotation of human, mouse and worm genes with mRNAs or ESTsAceView.". https://www.ncbi.nlm.nih.gov/ieb/research/acembly/av.cgi?db=human&term=C16orf71&submit=Go.
- ↑ "C16orf71 Symbol Report | HUGO Gene Nomenclature Committee". https://www.genenames.org/cgi-bin/gene_symbol_report?hgnc_id=HGNC:25081.
- ↑ "C16orf71 chromosome 16 open reading frame 71 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/146562.
- ↑ 7.0 7.1 "C16orf71 chromosome 16 open reading frame 71 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/?term=homo+sapiens+c16orf71.
- ↑ 8.0 8.1 "Transcript: C16orf71-003 (ENST00000586256.1) - Summary - Homo sapiens - Ensembl genome browser 88" (in en-gb). http://www.ensembl.org/Homo_sapiens/Transcript/Summary?db=core;g=ENSG00000166246;r=16:4734272-4749396;t=ENST00000586256.
- ↑ "PREDICTED: Homo sapiens chromosome 16 open reading frame 71 (C16orf71) - Nucleotide - NCBI". https://www.ncbi.nlm.nih.gov/nuccore/XM_017022976.1.
- ↑ "PREDICTED: Homo sapiens chromosome 16 open reading frame 71 (C16orf71) - Nucleotide - NCBI". https://www.ncbi.nlm.nih.gov/nuccore/XM_005255144.3.
- ↑ "PREDICTED: Homo sapiens chromosome 16 open reading frame 71 (C16orf71) - Nucleotide - NCBI". https://www.ncbi.nlm.nih.gov/nuccore/XM_017022975.1.
- ↑ "Cell atlas - C16orf71 - The Human Protein Atlas". http://www.proteinatlas.org/ENSG00000166246-C16orf71/cell#human.
- ↑ 13.0 13.1 13.2 13.3 "SDSC Biology Workbench". http://seqtool.sdsc.edu.
- ↑ "AMINO ACID FREQUENCY". http://www.tiem.utk.edu/~gross/bioed/webmodules/aminoacid.htm.
- ↑ Aungier, S. P. M.; Roche, J. F.; Duffy, P.; Scully, S.; Crowe, M. A. (2015-03-01). "The relationship between activity clusters detected by an automatic activity monitor and endocrine changes during the periestrous period in lactating dairy cows" (in English). Journal of Dairy Science 98 (3): 1666–1684. doi:10.3168/jds.2013-7405. ISSN 0022-0302. PMID 25529424. http://www.journalofdairyscience.org/article/S0022-0302(14)00871-6/fulltext.
- ↑ 16.0 16.1 Spector, David L.; Lamond, Angus I. (2011-02-01). "Nuclear Speckles" (in en). Cold Spring Harbor Perspectives in Biology 3 (2): a000646. doi:10.1101/cshperspect.a000646. ISSN 1943-0264. PMID 20926517.
- ↑ 17.0 17.1 "I-TASSER server for protein structure and function prediction". http://zhanglab.ccmb.med.umich.edu/I-TASSER/.
- ↑ "Redirecting to Phyre2" (in en-US). http://www.sbg.bio.ic.ac.uk/~phyre/.
- ↑ "NucPred - Home". http://www.sbc.su.se/~maccallr/nucpred/.
- ↑ 20.0 20.1 "GDS3688 / 222089_s_at". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS3688:222089_s_at.
- ↑ "GDS2761 / GI_21040258-S". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS2761:GI_21040258-S.
- ↑ 22.0 22.1 "GDS2696 / GI_21040258-S". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS2696:GI_21040258-S.
- ↑ "GDS2761 / GI_21040258-S". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS2761:GI_21040258-S.
- ↑ Semenza, Gregg (February 2012). "Hypoxia-Inducible Factors in Physiology and Medicine". Cell 148 (3): 399–408. doi:10.1016/j.cell.2012.01.021. PMID 22304911.
- ↑ "Home - EST - NCBI". https://www.ncbi.nlm.nih.gov/nucest/.
- ↑ "C16ORF71 - Chemical Interactions | CTD" (in en). http://ctd.mdibl.org/detail.go;jsessionid=EA79DEFE5F02834AC42F28491177DE69?type=gene&acc=146562&view=ixn.
- ↑ 27.0 27.1 27.2 Ali, Sazan; Steinmetz, Gérard; Montillet, Guillaume; Perrard, Marie-Hélène; Loundou, Anderson; Durand, Philippe; Guichaoua, Marie-Roberte; Prat, Odette (2014-09-02). "Exposure to Low-Dose Bisphenol A Impairs Meiosis in the Rat Seminiferous Tubule Culture Model: A Physiotoxicogenomic Approach". PLOS ONE 9 (9): e106245. doi:10.1371/journal.pone.0106245. ISSN 1932-6203. PMID 25181051. Bibcode: 2014PLoSO...9j6245A.
- ↑ 28.0 28.1 28.2 Song, Mi-Kyung; Lee, Hyo-Sun; Ryu, Jae-Chun (2015). "Integrated analysis of microRNA and mRNA expression profiles highlights aldehyde-induced inflammatory responses in cells relevant for lung toxicity". Toxicology 334: 111–121. doi:10.1016/j.tox.2015.06.007. PMID 26079696.
- ↑ 29.0 29.1 29.2 Tait, Sabrina; La Rocca, Cinzia; Mantovani, Alberto (2011-07-01). "Exposure of human fetal penile cells to different PCB mixtures: transcriptome analysis points to diverse modes of interference on external genitalia programming". Reproductive Toxicology 32 (1): 1–14. doi:10.1016/j.reprotox.2011.02.001. PMID 21334430.
- ↑ 30.0 30.1 30.2 30.3 "Genomatix - NGS Data Analysis & Personalized Medicine". http://www.genomatix.de/.
- ↑ "ZNF500 Gene". https://www.genecards.org/cgi-bin/carddisp.pl?gene=ZNF500.
- ↑ 32.0 32.1 32.2 32.3 "BLAST: Basic Local Alignment Search Tool". https://blast.ncbi.nlm.nih.gov/Blast.cgi.
- ↑ "Human BLAT Search". https://genome.ucsc.edu/cgi-bin/hgBlat.
 |


