Biology:Cas12a
| CRISPR-associated protein 12a | |
|---|---|
 | |
| Identifiers | |
| Symbol | Cas12a |
| InterPro | IPR027620 |
Template:CRISPR Cas12a (CRISPR-associated protein 12a, previously known as Cpf1) is an RNA-guided endonuclease-exonuclease that forms an essential component of the CRISPR systems found in some bacteria and archaea.[1] In its natural context, Cas12a targets and destroys the genetic material of viruses and other foreign mobile genetic elements, thereby protecting the host cell from infection. Like other Cas enzymes, Cas12a binds to a "guide" RNA (termed a crRNA, or CRISPR RNA) which targets it to a DNA sequence in a specific and programmable matter. In the host organism, the crRNA contains a constant region that is recognized by the Cas12a protein and a "spacer" region that is complementary to a piece of foreign nucleic acid (e.g. a portion of a phage genome) that previously infected the cell.[2]
As with Cas9 and other Cas proteins, the programmable DNA-targeting activity of Cas12a makes it a useful tool for biotechnology and biological research applications. By modifying the spacer sequence in the crRNA, researchers can target Cas12a to specific DNA sequences, allowing for highly targeted modifications of DNA.[3] Cas12a is distinguished from Cas9 by a its single RuvC endonuclease active site, its 5' protospacer adjacent motif preference, and its formation of sticky rather than blunt ends at the cut site; these and other differences may make it more suitable for certain applications . Beyond its use in basic research, CRISPR-Cas12a could have applications in the treatment of genetic illnesses and in implementing gene drives.[3]
History
Cas12a, a Class II Type V CRISPR-associated nuclease, was characterized in 2015 and was formerly known as Cpf1.[4] This nuclease is found in the CRISPR-Cpf1 system of bacteria such as Francisella novicida.[5][6] The initial designation, derived from a TIGRFAMs protein family definition established in 2012, reflected the prevalence of this CRISPR-Cas subtype in the Prevotella and Francisella lineages. Cas12a exhibits several key distinctions from Cas9: it generates staggered cuts in double-stranded DNA, in contrast to the blunt ends produced by Cas9;[7] it relies on a 'T-rich' protospacer adjacent motif (PAM) (typically 5'-TTTV-3', where V is A, C, or G), offering alternative targeting sites compared to the 'G-rich' PAMs (typically 5'-NGG-3') favored by Cas9;[8] and it requires only a CRISPR RNA (crRNA) for effective targeting, whereas Cas9 necessitates both a crRNA and a trans-activating crRNA (tracrRNA).[4]
These differences may confer certain advantages to Cas12a over Cas9. For instance, the smaller size of Cas12a's crRNAs makes them well-suited for multiplexed genome editing, allowing for a greater number of guide RNAs to be packaged into a single vector compared to Cas9's single guide RNAs (sgRNAs).[9] The sticky 5′ overhangs generated by Cas12a can also be utilized for DNA assembly with higher target specificity than traditional restriction enzyme cloning. Furthermore, Cas12a cleaves DNA 18–23 base pairs downstream from the PAM site. This characteristic ensures that the recognition sequence remains intact after repair, enabling Cas12a to perform multiple rounds of DNA cleavage. Conversely, Cas9 cleaves only 3 base pairs upstream of the PAM site, leading to indel mutations from the non-homologous end joining (NHEJ) pathway that can disrupt the recognition sequence, thereby preventing subsequent rounds of cutting. A notable feature of Cas12a, in contrast to Cas9, is its ability to remain bound to its target DNA after cleavage and subsequently cleave other single-stranded DNA molecules non-discriminately . This property, termed "collateral cleavage" or "trans-cleavage" activity, has been harnessed for the development of various diagnostic technologies.[4]
Comparison with Cas9
While classified as a Class II Type V CRISPR-associated nuclease, Cas12a lacks the NHN endonuclease domain found in Cas9 and instead relies on its RuvC-like domain for sequential DNA cleavage, resulting in the production of staggered ends.[4] Unlike Cas9, Cas12a possesses a unique zinc finger-like domain, which may contribute to DNA binding or structural stability. A significant advantage of Cas12a over Cas9 is its inherent RNase activity within the WED domain, enabling the self-processing of precursor crRNAs (pre-crRNAs) into mature crRNAs.[10] This eliminates the requirement for a trans-activating crRNA (tracrRNA), simplifying guide RNA design and facilitating multiplexed editing through the packaging of multiple crRNAs into a single vector.
Multiplex genome editing
Multiplex genome editing is a powerful technique that allows for the simultaneous editing of multiple genes. Cas12a is particularly useful in this context due to features that simplify its application compared to Cas9.[11] For instance, Cas12a requires only a short CRISPR RNA (crRNA) to target and edit genes.[12] These crRNAs can be easily multiplexed by packaging them together into a single delivery system, enabling the simultaneous editing of multiple genomic loci .[13]
Furthermore, Cas12a and Cas9 exhibit differences in their protospacer adjacent motif (PAM) requirements. While Cas9 typically looks for G-rich sequences, Cas12a targets T-rich sequences. This difference expands the range of targetable genomic sites, allowing Cas12a to edit genes in regions where Cas9 may not be efficient, such as AT-rich areas prevalent in plant genomes,[14] providing greater flexibility for researchers.
The ability of Cas12a to deliver its own crRNA (CRISPR RNA) without the need for additional components like the tracrRNA required by Cas9 further simplifies the design and delivery of guide RNAs for multiplexed editing.
Description
Discovery
CRISPR-Cas12a was found by searching a published database of bacterial genetic sequences for promising bits of DNA. Its identification through bioinformatics as a CRISPR system protein, its naming, and a hidden Markov model (HMM) for its detection were provided in 2012 in a release of the TIGRFAMs database of protein families. Cas12a appears in many bacterial species. The ultimate Cas12a endonuclease that was developed into a tool for genome editing was taken from one of the first 16 species known to harbor it.[15] Two candidate enzymes from Acidaminococcus and Lachnospiraceae display efficient genome-editing activity in human cells.[3]
Classification
CRISPR-Cas systems are separated into two classes: Class I, in which several Cas proteins associate with a crRNA to build a functional endonuclease, and Class II, in which a single Cas endonuclease associates with a crRNA; Class II is further divided into Type II, Type V, and Type VI systems. Cas12a is identified as a Class II, Type V CRISPR-Cas system.[16]
Naming
The acronym CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) refers to the invariant DNA sequences found in bacteria and archaea which encode Cas proteins and their crRNAs. Cas12a was originally known as Cpf1, an abbreviation of CRISPR and two genera of bacteria where it appears, Prevotella and Francisella. It was renamed in 2015 after a broader rationalization of the names of Cas (CRISPR associated) proteins to correspond to their sequence homology.[16]
Structure
The Cas12a protein contains a mixed alpha/beta domain, a RuvC-like endonuclease domain (broken into two non-contiguous segments, RuvC-I and RuvC-II) similar to the RuvC domain of Cas9, and a zinc finger-like domain.[17] Unlike Cas9, Cas12a does not have an HNH endonuclease domain, and the N-terminal region of Cas12a does not have an alpha-helical recognition lobe as seen in Cas9.[16]
The Cas12a loci encode Cas1, Cas2 and Cas4 proteins more similar to types I and III than from type II systems. Database searches suggest the abundance of Cas12a-family proteins in many bacterial species.[16]
Also unlike Cas9, Cas12a does not require a tracrRNA (which in natural CRISPR systems must base-pair with a separate crRNA before binding to a Cas protein), instead binding a single crRNA. Both Cas12a and its guide RNA are smaller than the protein and RNA components of the Cas9 system; the crRNA of Cas12a is approximately half as long as sgRNAs used with Cas9.[18] This reduced size renders Cas12a more suitable for applications such as in vivo delivery via adeno-associated virus (AAV), which have limited DNA packaging capacity due to their small capsids.
The Cas12a-crRNA complex cleaves target DNA or RNA by identification of a protospacer adjacent motif (PAM) 5'-YTN-3'[19] (where "Y" is a pyrimidine[20] and "N" is any nucleobase), in contrast to the G-rich PAM targeted by Cas9. After identification of PAM, Cas12a introduces a sticky-end-like DNA double- stranded break of 4 or 5 nucleotides overhang.[17]
Mechanism
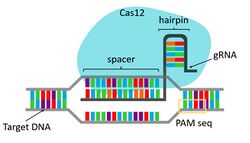
The CRISPR-Cas12a system consist of a Cas12a enzyme and a guide RNA that finds and positions the complex at the correct spot on the double helix to cleave target DNA. CRISPR-Cas12a systems activity has three stages:[15]
- Adaptation: Cas1 and Cas2 proteins facilitate the adaptation of small fragments of DNA into the CRISPR array.
- Formation of crRNAs: processing of pre-cr-RNAs producing of mature crRNAs to guide the Cas protein.
- Interference: the Cas12a is bound to a crRNA to form a binary complex to identify and cleave a target DNA sequence. The crRNA-Cas12a complex searches dsDNA for a 3-6nt 5' protospacer adjacent motif (PAM). Once a PAM is found, the protein locally denatures the dsDNA and searches for complementarity between the crRNA spacer and the ssDNA protospacer. Sufficient complementarity will trigger RuvC activity and the RuvC active site will then cut the non-target strand and then the target strand, ultimately generating a staggered dsDNA break with 5' ssDNA overhangs (cis cleavage).[17][21]
Cas9 vs. Cas12a
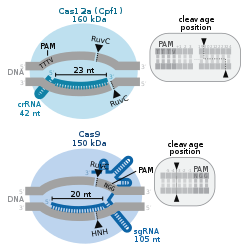
Cas9 requires two RNA molecules to cut DNA while Cas12a needs one. The proteins also cut DNA at different places, offering researchers more options when selecting an editing site. Cas9 cuts both strands in a DNA molecule at the same position, leaving behind blunt ends. Cas12a leaves one strand longer than the other, creating sticky ends. The sticky ends have different properties than blunt ends during non-homologous end joining or homologous repair of DNA, which confers certain advantages to Cas12a when attempting gene insertions, compared to Cas9.[15] Although the CRISPR-Cas9 system can efficiently disable genes, it is challenging to insert genes or generate a knock-in.[2] Cas12a lacks tracrRNA, utilizes a T-rich PAM and cleaves DNA via a staggered DNA DSB.[18]
In summary, important differences between Cas12a and Cas9 systems are that Cas12a:[22]
- Recognizes different PAMs, enabling new targeting possibilities.
- Creates 4-5 nt long sticky ends, instead of blunt ends produced by Cas9, enhancing the efficiency of genetic insertions and specificity during NHEJ or HDR.
- Cuts target DNA further away from PAM, further away from the Cas9 cutting site, enabling new possibilities for cleaving the DNA.
| Feature | Cas9 | Cas12a |
|---|---|---|
| Structure | Two RNA required (Or 1 fusion transcript (crRNA+tracrRNA=sgRNA) | One crRNA required |
| Cutting mechanism | Blunt end cuts | Staggered end cuts |
| Cutting site | Proximal to recognition site | Distal from recognition site |
| Target sites | G-rich PAM | T-rich PAM |
Origin
Cas12 endonucleases ultimately likely evolved from the TnpB endonuclease of IS200/IS605-family transposons. TnpB, not yet "domesticated" into the CRISPR immune system, are themselves able to perform RNA-guided cleavage using a OmegaRNA template system.[23]
Tools
Multiple aspects influence target efficiency and specificity when using CRISPR, including guide RNA design. Many design models and CRISPR-Cas software tools for optimal design of guide RNA have been developed. These include SgRNA designer, CRISPR MultiTargeter, SSFinder.[24] In addition, commercial antibodies are available for use to detect Cas12a protein.[25]
Intellectual property
CRISPR-Cas9 is subject to Intellectual property disputes while CRISPR-Cas12a does not have the same issues.[3]
Notes
References
- ↑ Bhattacharya, Supreet; Agarwal, Ankit; Muniyappa, Kalappa (2024). "Deciphering the Substrate Specificity Reveals that CRISPR-Cas12a is a Bifunctional Enzyme with Both Endo- and Exonuclease Activities". Journal of Molecular Biology 436 (10). doi:10.1016/j.jmb.2024.168550. PMID 38575054.
- ↑ 2.0 2.1 "CRISPR-Based Genetic Engineering Gets a Kick in the Cas" (in en-US). 2015-09-29. http://news.meta.com/?p=863.
- ↑ 3.0 3.1 3.2 3.3 "Even CRISPR". The Economist. ISSN 0013-0613. https://www.economist.com/news/science-and-technology/21668031-scientists-have-found-yet-another-way-edit-genomes-suggesting-such-technology-will.
- ↑ 4.0 4.1 4.2 4.3 "The CRISPR-Cas immune system: biology, mechanisms and applications". Biochimie 117: 119–128. October 2015. doi:10.1016/j.biochi.2015.03.025. PMID 25868999.
- ↑ "What is CRISPR/Cas9?". Archives of Disease in Childhood: Education and Practice Edition 101 (4): 213–215. August 2016. doi:10.1136/archdischild-2016-310459. PMID 27059283.
- ↑ "CRISPR/Cas, the immune system of bacteria and archaea". Science 327 (5962): 167–170. January 2010. doi:10.1126/science.1179555. PMID 20056882. Bibcode: 2010Sci...327..167H.
- ↑ CRISPR-CAS9, TALENS and ZFNS – the battle in gene editing https://www.ptglab.com/news/blog/crispr-cas9-talens-and-zfns-the-battle-in-gene-editing/
- ↑ "Press release: The Nobel Prize in Chemistry 2020". Nobel Foundation. https://www.nobelprize.org/prizes/chemistry/2020/press-release/.
- ↑ "Cysteine proteases: Battling pathogenic parasitic protozoans with omnipresent enzymes". Microbiological Research 249. August 2021. doi:10.1016/j.micres.2021.126784. PMID 33989978.
- ↑ The Code Breaker: Jennifer Doudna, Gene Editing, and the Future of the Human Race. New York: Simon & Schuster. 2021. p. 73. ISBN 978-1-9821-1585-2. OCLC 1239982737. https://books.google.com/books?id=f_D3DwAAQBAJ. Retrieved 2021-10-20.
- ↑ CRISPR-Cas Systems: RNA-mediated Adaptive Immunity in Bacteria and Archaea. Heidelberg: Springer. 2013. p. 6. ISBN 978-3-642-34656-9.
- ↑ "Small CRISPR RNAs guide antiviral defense in prokaryotes". Science 321 (5891): 960–964. August 2008. doi:10.1126/science.1159689. PMID 18703739. Bibcode: 2008Sci...321..960B.
- ↑ "Candida albicans by Increased Single Guide RNA Expression". mSphere 2 (2): e00385–16. 2017. doi:10.1128/mSphere.00385-16. PMID 28435892.
- ↑ "Efficient genome editing in zebrafish using a CRISPR-Cas system". Nature Biotechnology 31 (3): 227–229. March 2013. doi:10.1038/nbt.2501. PMID 23360964.
- ↑ 15.0 15.1 15.2 "Bacteria yield new gene cutter". Nature 526 (7571): 17. October 2015. doi:10.1038/nature.2015.18432. PMID 26432219.
- ↑ 16.0 16.1 16.2 16.3 "An updated evolutionary classification of CRISPR-Cas systems". Nature Reviews. Microbiology 13 (11): 722–736. November 2015. doi:10.1038/nrmicro3569. PMID 26411297.
- ↑ 17.0 17.1 17.2 "Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system". Cell 163 (3): 759–771. October 2015. doi:10.1016/j.cell.2015.09.038. PMID 26422227.
- ↑ 18.0 18.1 "Cpf1 Moves in on Cas9 for Next-Gen CRISPR Genome Editing". 29 September 2015. http://epigenie.com/cpf1-takes-crispr-bigger-by-going-smaller/.
- ↑ "The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA". Nature 532 (7600): 517–521. April 2016. doi:10.1038/nature17945. PMID 27096362. Bibcode: 2016Natur.532..517F.
- ↑ "Nucleotide Codes, Amino Acid Codes, and Genetic Codes". KEGG: Kyoto Encyclopedia of Genes and Genomes. July 15, 2014. http://www.genome.jp/kegg/catalog/codes1.html.
- ↑ "CRISPR-Cas12a exploits R-loop asymmetry to form double-strand breaks". eLife 9. June 2020. doi:10.7554/eLife.55143. PMID 32519675.
- ↑ "Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA". Cell 165 (4): 949–962. May 2016. doi:10.1016/j.cell.2016.04.003. PMID 27114038.
- ↑ "The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases". Science 374 (6563): 57–65. October 2021. doi:10.1126/science.abj6856. PMID 34591643. Bibcode: 2021Sci...374...57A.
- ↑ "Resources for the design of CRISPR gene editing experiments". Genome Biology 16: 260. November 2015. doi:10.1186/s13059-015-0823-x. PMID 26612492.
- ↑ "Anti-CPF1 antibody (GTX133301) | GeneTex". https://www.genetex.com/Product/Detail/CPF1-antibody/GTX133301.
 |
