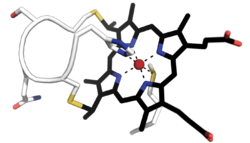
Biology:Cytochrome c family
Cytochromes c (cyt c, c-type cytochromes) cytochromes, or heme-containing proteins, that have heme C covalently attached to the peptide backbone via one or two thioether bonds.[1] These bonds are in most cases part of a specific Cys-X-X-Cys-His (CXXCH) binding motif, where X denotes a miscellaneous amino acid. Two thioether bonds of cysteine residues bind to the vinyl sidechains of heme, and the histidine residue coordinates one axial binding site of the heme iron. Less common binding motifs can include a single thioether linkage,[2] a lysine[3] or a methionine[4] instead of the axial histidine or a CXnCH binding motif with n>2.[5] The second axial site of the iron can be coordinated by amino acids of the protein,[6] substrate molecules or water. Cytochromes c possess a wide range of properties and function as electron transfer proteins or catalyse chemical reactions involving redox processes.[7] A prominent member of this family is mitochondrial cytochrome c.
Classification
| Cytochrome c (Class I) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
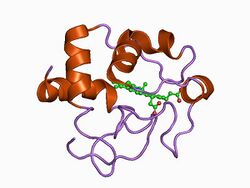 | |||||||||
| Identifiers | |||||||||
| Symbol | Cytochrom_C | ||||||||
| Pfam | PF00034 | ||||||||
| InterPro | IPR009056 | ||||||||
| PROSITE | PDOC00169 | ||||||||
| SCOP2 | 1cry / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 71 | ||||||||
| OPM protein | 1hrc | ||||||||
| Membranome | 210 | ||||||||
| |||||||||
| Cytochrome c (Class II) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
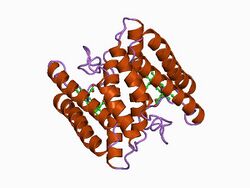 | |||||||||
| Identifiers | |||||||||
| Symbol | Cytochrom_C_2 | ||||||||
| Pfam | PF01322 | ||||||||
| InterPro | IPR002321 | ||||||||
| PROSITE | PDOC00169 | ||||||||
| SCOP2 | 1cgo / SCOPe / SUPFAM | ||||||||
| |||||||||
| High molecular weight cytochrome c (Class III) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of the 16-heme cytochrome c Hmc from Desulfovibrio vulgaris hildenborough (PDB: 1H29; P24092). | |||||||||
| Identifiers | |||||||||
| Symbol | Cytochrom_CIII | ||||||||
| Pfam | PF02085 | ||||||||
| Pfam clan | CL0317 | ||||||||
| InterPro | IPR020942 | ||||||||
| SCOP2 | 2cdv / SCOPe / SUPFAM | ||||||||
| CDD | cd08168 | ||||||||
| |||||||||
Cytochrome c proteins can be divided in four classes based on their size, number of heme groups and reduction potentials:[9]
Class I
Small soluble cytochrome c proteins with a molecular weight of 8-12 kDa and a single heme group belong to class I.[10][11] It includes the low-spin soluble cytC of mitochondria and bacteria, with the heme-attachment site located towards the N-terminus, and the sixth ligand provided by a methionine residue about 40 residues further on towards the C-terminus. The typical class I fold contains five α-helices. On the basis of sequence similarity, class I cytC were further subdivided into five classes, IA to IE. Class IB includes the eukaryotic mitochondrial cyt c and prokaryotic 'short' cyt c2 exemplified by Rhodopila globiformis cyt c2; class IA includes 'long' cyt c2, such as Rhodospirillum rubrum cyt c2 and Aquaspirillum itersonii cyt c550, which have several extra loops by comparison with class IB cyt c.
The linked InterPro entry represents mono-haem cytochrome c proteins (excluding class II and f-type cytochromes), such as cytochromes c, c1, c2, c5, c555, c550-c553, c556, c6 and cbb3. Diheme cytochrome c (InterPro: IPR018588) are proteins with a class I cluster and a unique cluster.
Subclasses
- Cytochrome c, class IA/IB InterPro: IPR002327
- Cytochrome c, class IC InterPro: IPR008168
- Cytochrome c, class ID InterPro: IPR002324
- Cytochrome c, class IE InterPro: IPR002323
Class II
The heme group in class II cytochrome c proteins is attached to a C-terminal binding motif. The structural fold of class II c-type cytochromes contains a four α-helix bundle with the covalently attached heme group at its core.[12] Representatives of class II are the high-spin cytochrome c' and a number of low-spin cytochromes c, e.g. cyt c556. The cyt c' are capable of binding such ligands as CO, NO or CN−, albeit with rate and equilibrium constants 100 to 1,000,000-fold smaller than other high-spin hemeproteins.[13] This, coupled with its relatively low redox potential, makes it unlikely that cyt c' is a terminal oxidase. Thus cyt c' probably functions as an electron transfer protein.[12] The 3D structures of a number of cyt c' have been determined which show that the proteins usually exist as a dimer. The Chromatium vinosum cyt c' exhibits dimer dissociation upon ligand binding.[14]
Class III
Proteins containing multiple covalently attached heme groups with low redox potential are included in class III. The heme C groups, all bis-histidinyl coordinated, are structurally and functionally nonequivalent and present different redox potentials in the range 0 to -400 mV.[15] Members of this class are e.g. cytochrome c7 (triheme), cytochrome c3 (tetraheme), and high-molecular-weight cytochrome c (Hmc), containing 16 heme groups with only 30-40 residues per heme group.[16] The 3D structures of a number of cyt c3 proteins have been determined. The proteins consist of 4-5 α-helices and 2 β-sheets wrapped around a compact core of four non-parallel hemes, which present a relatively high degree of exposure to the solvent. The overall protein architecture, heme plane orientations and iron-iron distances are highly conserved.[15]
An example is the Photosynthetic reaction centre of Rhodopseudomonas viridis that contains a tetraheme cytochrome c subunit.[17]
Class IV
According to Ambler (1991), Cytochrome c proteins containing other prosthetic groups besides heme C, such as flavocytochromes c (sulfide dehydrogenase) and cytochromes cd1 (nitrite reductase) belong to class IV.[9] As this grouping is more related to how the heme group is used instead of what the domains themselves look like, proteins placed in this group tend to be scattered in others in bioinformatic groupings.
Biogenesis
The attachment of the heme group is physically separated from the protein biosynthesis. Proteins are synthesized within the cytoplasm and endoplasmic reticulum, while the maturation of cytochromes c occurs in the periplasm of prokaryots, the intermembrane space of mitochondria or the stroma of chloroplasts. Several biochemical pathways have been discovered that differ depending on organism.[18]
System I
Also called cytochrome c maturation (ccm) and found in Pseudomonadota, plant mitochondria, some protozoal mitochondria, deinococci, and archaea.[19] Ccm comprises at least eight membrane proteins (CcmABCDEFGH) that are needed for electron transfer to the heme group, apo-cytochrome handling and attachment of the heme to the apo-cytochrome. An ABC-transporter-like complex formed by CcmA2BCD attaches a heme group to CcmE with the use of ATP. CcmE transports the heme to CcmF where the attachment to the apo-cytochrome occurs. Transport of the apoprotein from the cytoplasm to the periplasm happens via the Sec translocation system. CcmH is used by the system to recognize the apo-cytochrome and direct it to CcmF.
System II
Cytochromes c in chloroplasts, Gram-positive bacteria, cyanobacteria, and some Pseudomonadota are produced by the cytochrome c synthesis (ccs) system. It is composed of two membrane proteins CcsB and CcsA. The CcsBA protein complex was suggested to act as a heme transporter during the attachment process.[20] In some organisms such as Helicobacter hepaticus both proteins are found as a fused single protein. Apoprotein transport occurs via the Sec translocon as well.
System III
Fungal, vertebrate and invertebrate mitochondria produce cytochrome c proteins with a single enzyme called HCCS (holocytochrome c synthase) or cytochrome c heme lyase (CCHL).[21][22] The protein is attached to the inner membrane of the intermembrane space.[23] In some organisms, such as Saccharomyces cerevisiae, cytochrome c and cytochrome c1 are synthesized by separate heme lyases, CCHL and CC1HL respectively.[24] In Homo sapiens a single HCCS is used for the biosynthesis of both cytochrome c proteins.[25]
System IV
Four membrane proteins are necessary for the attachment of a heme in cytochrome b6. A major difference to systems I-III is that the heme attachment occurs at the opposite side of the lipid bilayer compared to the other systems.[18]
Human proteins containing this domain
References
- ↑ "Nomenclature Committee of the International Union of Biochemistry (NC-IUB). Nomenclature of electron-transfer proteins. Recommendations 1989". The Journal of Biological Chemistry 267 (1): 665–77. January 1992. doi:10.1016/S0021-9258(18)48544-4. PMID 1309757.
- ↑ "Maturation of the unusual single-cysteine (XXXCH) mitochondrial c-type cytochromes found in trypanosomatids must occur through a novel biogenesis pathway". The Biochemical Journal 383 (Pt. 3): 537–42. November 2004. doi:10.1042/BJ20040832. PMID 15500440.
- ↑ "Involvement of products of the nrfEFG genes in the covalent attachment of haem c to a novel cysteine-lysine motif in the cytochrome c552 nitrite reductase from Escherichia coli". Molecular Microbiology 28 (1): 205–16. April 1998. doi:10.1046/j.1365-2958.1998.00792.x. PMID 9593308.
- ↑ "X-ray structure of the membrane-bound cytochrome c quinol dehydrogenase NrfH reveals novel haem coordination". The EMBO Journal 25 (24): 5951–60. December 2006. doi:10.1038/sj.emboj.7601439. PMID 17139260.
- ↑ "A dedicated haem lyase is required for the maturation of a novel bacterial cytochrome c with unconventional covalent haem binding". Molecular Microbiology 64 (4): 1049–60. May 2007. doi:10.1111/j.1365-2958.2007.05712.x. PMID 17501927. https://ueaeprints.uea.ac.uk/31288/1/Hartshorne_MCCA.pdf.
- ↑ "Structural model for an alkaline form of ferricytochrome C". Journal of the American Chemical Society 125 (10): 2913–22. March 2003. doi:10.1021/ja027180s. PMID 12617658. https://authors.library.caltech.edu/76575/2/ja027180ssi20021029_013138%20%282%29.pdf.
- ↑ Pettigrew, Graham W.; Moore, Geoffrey R. (1987). "The Function of Bacterial and Photosynthetic Cytochromes C". Cytochromes C. Springer Series in Molecular Biology. Berlin Heidelberg: Springer. pp. 113–229. doi:10.1007/978-3-642-72698-9_3. ISBN 978-3-642-72698-9. https://archive.org/details/cytochromescbiol0000pett.
- ↑ "Application of an automatic molecular-replacement procedure to crystal structure analysis of cytochrome c2 from Rhodopseudomonas viridis". Acta Crystallogr. D 50 (Pt 3): 271–5. May 1994. doi:10.1107/S0907444993013952. PMID 15299438.
- ↑ 9.0 9.1 "Sequence variability in bacterial cytochromes c". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1058 (1): 42–7. May 1991. doi:10.1016/S0005-2728(05)80266-X. PMID 1646017.
- ↑ "Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers" (in EN). Chemical Reviews 114 (8): 4366–469. April 2014. doi:10.1021/cr400479b. PMID 24758379.
- ↑ "Multifunctional Cytochrome c: Learning New Tricks from an Old Dog". Chemical Reviews 117 (21): 13382–13460. November 2017. doi:10.1021/acs.chemrev.7b00257. PMID 29027792.
- ↑ 12.0 12.1 "Bacterial 4-alpha-helical bundle cytochromes". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1058 (1): 38–41. May 1991. doi:10.1016/s0005-2728(05)80265-8. PMID 1646016.
- ↑ "Ligand binding properties of cytochromes c'". Biochimica et Biophysica Acta 1058 (1): 8–12. May 1991. doi:10.1016/s0005-2728(05)80257-9. PMID 1646027.
- ↑ "Atomic structure of a cytochrome c' with an unusual ligand-controlled dimer dissociation at 1.8 A resolution". Journal of Molecular Biology 234 (2): 433–45. November 1993. doi:10.1006/jmbi.1993.1597. PMID 8230224.
- ↑ 15.0 15.1 "[9] Tetraheme cytochromes". Tetraheme cytochromes. Methods in Enzymology. 243. 1994. pp. 119–40. doi:10.1016/0076-6879(94)43011-X. ISBN 9780121821449.
- ↑ "The crystal structure of the hexadeca-heme cytochrome Hmc and a structural model of its complex with cytochrome c(3)". Structure 10 (12): 1677–86. December 2002. doi:10.1016/s0969-2126(02)00909-7. PMID 12467575.
- ↑ "A comparison of stigmatellin conformations, free and bound to the photosynthetic reaction center and the cytochrome bc1 complex". Journal of Molecular Biology 368 (1): 197–208. April 2007. doi:10.1016/j.jmb.2007.02.013. PMID 17337272.
- ↑ 18.0 18.1 "Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control". Microbiology and Molecular Biology Reviews 73 (3): 510–28, Table of Contents. September 2009. doi:10.1128/MMBR.00001-09. PMID 19721088.
- ↑ "Cytochrome c biogenesis System I". The FEBS Journal 278 (22): 4170–8. November 2011. doi:10.1111/j.1742-4658.2011.08376.x. PMID 21958041.
- ↑ "CcsBA is a cytochrome c synthetase that also functions in heme transport". Proceedings of the National Academy of Sciences of the United States of America 106 (25): 10201–6. June 2009. doi:10.1073/pnas.0903132106. PMID 19509336.
- ↑ "Identification and sequence of the gene encoding cytochrome c heme lyase in the yeast Saccharomyces cerevisiae". The EMBO Journal 6 (1): 235–41. January 1987. doi:10.1002/j.1460-2075.1987.tb04744.x. PMID 3034577.
- ↑ "Biochemical requirements for the maturation of mitochondrial c-type cytochromes". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1793 (1): 125–38. January 2009. doi:10.1016/j.bbamcr.2008.06.017. PMID 18655808.
- ↑ "Mitochondrial cytochrome c biogenesis: no longer an enigma". Trends in Biochemical Sciences 40 (8): 446–55. August 2015. doi:10.1016/j.tibs.2015.05.006. PMID 26073510.
- ↑ "Biogenesis of mitochondrial heme lyases in yeast. Import and folding in the intermembrane space". The Journal of Biological Chemistry 270 (39): 22842–9. September 1995. doi:10.1074/jbc.270.39.22842. PMID 7559417.
- ↑ "Overlapping specificities of the mitochondrial cytochrome c and c1 heme lyases". The Journal of Biological Chemistry 278 (50): 49732–42. December 2003. doi:10.1074/jbc.M308881200. PMID 14514677.
 |