Biology:ERICH2
 Generic protein structure example |
Glutamate Rich Protein 2 is a protein in humans encoded by the gene ERICH2. This protein is expressed heavily in male tissues specifically in the testes, and proteins are specifically found in the nucleoli fibrillar center and the vesicles of these testicular cells.[1] The protein has multiple protein interactions which indicate that it may play a role in histone modification and proper histone functioning.[2]
Gene
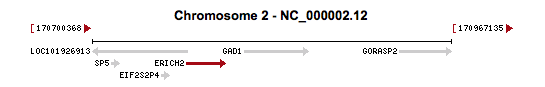
ERICH2 is located on human Chromosome 2, at 2q31.1.[3] It contains 10 distinct exons. The gene itself is 28,930 base pairs long and is flanked by the EIF2S2P4 and GAD1 genes.[3] There are no known paralogs of the ERICH2 gene.
mRNA
ERICH2 transcription produces three validated distinct mRNA variants. The longest transcript variant is 1,388 base pairs in length, 1,311 of which are coding.[3] The second variant differs from the first in its 5' UTR. It also has coding sequence differences and a distinct N-terminus compared to variant 1.[3] Variant 3 lacks several exons, has a distinct 3' UTR and C- terminus coding region. This variant is also shorter than the other two at 1,063 base pairs.[3]

Protein
The ERICH2 protein is 436 amino acids in length, and has a molecular weight of approximately 48,000 kD,[4] with an isoelectric point of approximately 5.[4] The protein is determined to be rich in the amino acid proline and low in tyrosine and glycine.
Motifs and domains

Two known motifs were found in the human ERICH2 protein. The KKNT motif functions in cAMP- and cGMP- dependent protein phosphorylation, this protein motif was found only in primates.[5] There is also a FGRR motif conserved in mammals that is defined as an amidation site.[6] Finally the ERICH2 protein contains the PHA03247 domain that is 32 amino acids long.[3] This domain is not generally conserved through orthologs and the function is unknown. It is present in the proteins that make up the herpes virion.[7]
Structure and localization
Secondary structure prediction shows one alpha helix and one beta strand formation. The alpha helix encompasses the entire conserved section as seen in the cartoon of the ERICH2 protein. The beta strand is predicted 12 amino acids down from the amidation site and encompasses 4 amino acids.[8] Four nuclear localization signals were found in the protein, two pat4 signals and two pat7 signals, their locations are shown in the cartoon.[9] It is predicted in the 78th percent that the protein resides in the nucleus.[9]
Expression

ERICH2 is not ubiquitously expressed. It however, has been shown to be expressed narrowly in the choroid plexus of a developing fetus and in the testes of adults.[11] Lung and female tissue expression were also present but expression was greatly decreased.[12] Proteins are specifically located in the nucleoli fibrillar center and the vesicles within cells.[1][13]
Regulation of expression
Many phosphorylation sites are predicted for the ERICH2 protein. None are predicted on tyrosines only on serines and the threonines.[14][15] There is also a predicted acetylation site at the N-terminus of the protein, specifically it is predicted on the third amino acid.[16] Many SOX/SRY-sex/testis determining and related HMG box factor transcription factors and estrogen related transcription factors are predicted to bind and regulate transcription of ERICH2.[17]
Function
Interacting proteins
ERICH2 interacts with proteins in the H2A family.[2][13] The H2A proteins specifically play a role in the octamer structure of histone. ERICH2 is specifically known to interact with the H2AFY protein, which plays a key role in the stable X chromosome inactivation and can function by replacing a normal H2A in certain nucleosomes and thus repressing transcription.[18]
ERICH2 is also known to interact with the protein SDCB1 which functions in vesicle trafficking and the regulation of growth and proliferation of certain cancer cells.[19]
The IWS1 protein also interacts with ERICH2. This protein functions as a transcription factor and plays a key role in defining the composition of the RNA polymerase II elongation complex.[20] This complex then plays a role in histone modification and proper splicing.
Two-hybrid assays and other protein interaction methods have shown an interaction with the PSORS1C2 protein, but the function of this protein remains unknown.[2]
Homology
No paralogs for the ERICH2 protein are known. ERICH2 has 124 known orthologs spanning multiple taxa.[3]
| Genus and Species | Common Name | Date of Divergence (MYA)[21] | Sequence Length (aa) | Sequence Identity | Sequence similarity |
| Homo sapiens | Human | 0 | 436 | -- | -- |
| Rousettus aegyptiacus | Egyptian fruit bat | 94 | 430 | 58% | 63% |
| Propithecus coquereli | Coquerel's sifaka | 74 | 323 | 54% | 60% |
| Mus musculus | Mouse | 90 | 463 | 47% | 56% |
| Ursus Maritimus | Polar Bear | 94 | 296 | 50% | 53% |
| Alligator mississippiensis | American Alligator | 320 | 370 | 28% | 38% |
| Thamnophis sirtalis | Common Garter Snake | 320 | 309 | 27% | 37% |
| Callorhinchus milii | Australian Ghost Shark | 465 | 319 | 22% | 33% |
| Danio rerio | Zebra Fish | 432 | 310 | 24% | 30% |
| Strongylocentrotus purpuratus | Purple Sea Urchin | 627 | 470 | 23% | 25% |
| Crassostrea gigas | Pacific Oyster | 758 | 293 | 17% | 22% |
| Bemisia tabaci | Silverleaf Whitefly | 758 | 213 | 13% | 14% |
| Trichoplax adhaerens | Trichoplax | 930 | 164 | 12% | 15% |
References
- ↑ 1.0 1.1 "Cell atlas - ERICH2 - The Human Protein Atlas". http://www.proteinatlas.org/ENSG00000204334-ERICH2/cell.
- ↑ 2.0 2.1 2.2 "Results - mentha: the interactome browser". http://mentha.uniroma2.it/result.php#erich2.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 "ERICH2 glutamate rich 2 [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/285141.
- ↑ 4.0 4.1 Kramer, Jack (1990). "Biology WorkBench 3.2". http://seqtool.sdsc.edu/CGI/BW.cgi#!.[yes|permanent dead link|dead link}}]
- ↑ "PROSITE" (in en-US). http://prosite.expasy.org/PS00005.
- ↑ "PROSITE" (in en-US). http://prosite.expasy.org/PS00009.
- ↑ "Genomic and phylogenetic evidence of VIPER retrotransposon domestication in trypanosomatids". Memórias do Instituto Oswaldo Cruz 111 (12): 765–769. December 2016. doi:10.1590/0074-02760160224. PMID 27849219.
- ↑ Pearson, William (1999). "Biology workbench". http://seqtool.sdsc.edu/CGI/BW.cgi#!.[yes|permanent dead link|dead link}}]
- ↑ 9.0 9.1 "PSORT II Prediction". https://psort.hgc.jp/form2.html.
- ↑ "Cell atlas - ERICH2 - The Human Protein Atlas". http://www.proteinatlas.org/ENSG00000204334-ERICH2/cell.
- ↑ European Molecular Biology Lab. "Expression Atlas". https://www.ebi.ac.uk/gxa/query?geneQuery=%5B%7B%22value%22%3A%22ERICH2%22%2C%22category%22%3A%22symbol%22%7D%5D&organism=&conditionQuery=%5B%5D&bs=%7B%22homo%20sapiens%22%3A%5B%22ORGANISM_PART%22%5D%2C%22bos%20taurus%22%3A%5B%22ORGANISM_PART%22%5D%2C%22chlorocebus%20sabaeus%22%3A%5B%22ORGANISM_PART%22%5D%2C%22equus%20caballus%22%3A%5B%22ORGANISM_PART%22%5D%2C%22macaca%20mulatta%22%3A%5B%22ORGANISM_PART%22%5D%2C%22mus%20musculus%22%3A%5B%22ORGANISM_PART%22%5D%2C%22ovis%20aries%22%3A%5B%22ORGANISM_PART%22%5D%2C%22papio%20anubis%22%3A%5B%22ORGANISM_PART%22%5D%2C%22rattus%20norvegicus%22%3A%5B%22ORGANISM_PART%22%5D%2C%22xenopus%20tropicalis%22%3A%5B%22ORGANISM_PART%22%5D%7D&ds=%7B%22kingdom%22%3A%5B%22animals%22%5D%7D#baseline.
- ↑ Group, Schuler. "EST Profile - Hs.443729". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.443729.
- ↑ 13.0 13.1 Database, GeneCards Human Gene. "ERICH2 Gene - GeneCards | ERIC2 Protein | ERIC2 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=ERICH2.
- ↑ "ExPASy: SIB Bioinformatics Resource Portal - Categories" (in en-US). https://www.expasy.org/proteomics/families__patterns_and_profiles.
- ↑ "NetPhos 3.1 Server" (in en). http://www.cbs.dtu.dk/services/NetPhos/.
- ↑ "NetAcet 1.0 Server" (in en). http://www.cbs.dtu.dk/services/NetAcet/.
- ↑ "Genomatix: Genome Annotation and Browser: Query Input" (in en-US). https://www.genomatix.de/cgi-bin/eldorado/eldorado.pl?s=6089e70074f8817e0a2fd54f442219ad.
- ↑ "H2AFY - Core histone macro-H2A.1 - Homo sapiens (Human) - H2AFY gene & protein" (in en). https://www.uniprot.org/uniprot/O75367.
- ↑ "SDCBP - Syntenin-1 - Homo sapiens (Human) - SDCBP gene & protein" (in en). https://www.uniprot.org/uniprot/O00560.
- ↑ Database, GeneCards Human Gene. "IWS1 Gene - GeneCards | IWS1 Protein | IWS1 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=IWS1.
- ↑ "TimeTree :: The Timescale of Life". http://www.timetree.org.
 |
