Biology:Phytoene desaturase (lycopene-forming)
| Phytoene desaturase (lycopene-forming) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystallographic structure of a bacterial phytoene desaturase monomer from Pantoea ananatis.[1] | |||||||||
| Identifiers | |||||||||
| EC number | 1.3.99.31 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
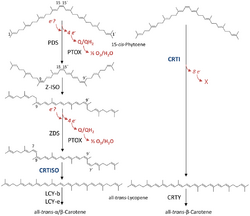
Phytoene desaturase (lycopene-forming) (CrtI, four-step phytoene desaturase) (EC 1.3.99.31, 15-cis-phytoene:acceptor oxidoreductase (lycopene-forming)) are enzymes found in archaea, bacteria and fungi that are involved in carotenoid biosynthesis.[2] They catalyze the conversion of colorless 15-cis-phytoene into a bright red lycopene in a biochemical pathway called the poly-trans pathway. The same process in plants and cyanobacteria utilizes four separate enzymes in a poly-cis pathway.[3]
Biochemistry
Bacterial phytoene desaturases were shown to require FAD as a cofactor for their function.[4] During the chemical reaction in total four additional double bonds are introduced into phytoene:
- 15-cis-phytoene + 4 acceptor [math]\displaystyle{ \rightleftharpoons }[/math] all-trans-lycopene + 4 reduced acceptor (overall reaction)
- (1a) 15-cis-phytoene + acceptor [math]\displaystyle{ \rightleftharpoons }[/math] all-trans-phytofluene + reduced acceptor
- (1b) all-trans-phytofluene + acceptor [math]\displaystyle{ \rightleftharpoons }[/math] all-trans-zeta-carotene + reduced acceptor
- (1c) all-trans-zeta-carotene + acceptor [math]\displaystyle{ \rightleftharpoons }[/math] all-trans-neurosporene + reduced acceptor:
- (1d) all-trans-neurosporene + acceptor [math]\displaystyle{ \rightleftharpoons }[/math] all-trans-lycopene + reduced acceptor
Applications
In 2000 it was discovered that the gene insertion of a bacterial phytoene desaturase into transgenic tomatoes increased the lycopene content without the need to alter several of the plants enzymes.[5] This approach was later used in rice to increase its β-carotene content resulting in the Golden Rice project.
See also
- 15-Cis-phytoene desaturase
- Phytoene desaturase (neurosporene-forming)
- Phytoene desaturase (zeta-carotene-forming)
- Phytoene desaturase (3,4-didehydrolycopene-forming)
References
- ↑ PDB: 4dgk; Schaub P; Yu Q; Gemmecker S; Poussin-Courmontagne P; Mailliot J; McEwen AG; Ghisla S; Al-Babili S et al. (2012). "On the Structure and Function of the Phytoene Desaturase CRTI from Pantoea ananatis, a Membrane-Peripheral and FAD-Dependent Oxidase/Isomerase". PLOS ONE 7 (6): e39550. doi:10.1371/journal.pone.0039550. PMID 22745782.
- ↑ Fraser, P.D.; Misawa, N.; Linden, H.; Yamano, S.; Kobayashi, K.; Sandmann, G. (1992). "Expression in Escherichia coli, purification, and reactivation of the recombinant Erwinia uredovora phytoene desaturase". J. Biol. Chem. 267 (28): 19891–19895. doi:10.1016/S0021-9258(19)88639-8. PMID 1400305.
- ↑ "Mechanistic aspects of carotenoid biosynthesis.". Chemical Reviews 114 (1): 164–193. 31 October 2013. doi:10.1021/cr400106y. PMID 24175570.
- ↑ Dailey TA; Dailey HA (May 1998). "Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases, and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus.". Journal of Biological Chemistry 273 (22): 13658–13662. doi:10.1074/jbc.273.22.13658. PMID 9593705.
- ↑ Romer, S.; Fraser, P.D.; Kiano, J.W.; Shipton, C.A.; Misawa, N; Schuch, W.; Bramley, P.M. (2000). "Elevation of provitamin A content of transgenic tomato plants.". Nature Biotechnology 18 (6): 666–669. doi:10.1038/76523. PMID 10835607.
External links
- Phytoene+desaturase+(lycopene-forming) at the US National Library of Medicine Medical Subject Headings (MeSH)
 |