Chemistry:Phenol sulfur transferase deficiency
Phenol sulfur transferase deficiency, in short PST deficiency, is the lack or the reduced activity of the functional enzyme phenol sulfur transferase, which is crucial in the detoxification of mainly phenolic compounds by catalysing the sulfate conjugation of the hydroxyl groups in the toxic phenolic compounds to result in more hydrophilic forms for more efficient excretion. This metabolic disorder was first discovered in the late 1990s by Dr. Rosemary Waring during her researches with autistic children, which also made this deficiency commonly associated to the topics of autism. Mutations in the PST genes account for the genetic causes of the deficiency, of which single nucleotide polymorphism and methylation of promoters are two examples of mutations that respectively cause conformational abnormalities and diminished expressions to the enzyme, resulting in the reduced detoxification of phenolic compounds and regulation of phenolic neurotransmitter. The deficiency may cause symptoms like flushing, tachycardia, and depression, and be a risk factor for disorders like autism, migraine, and cancer, while it also limits the use of phenolic drugs in PST deficient patients. There is currently no drug available for treating PST deficiency. However, some people suffering from PST deficiency have found taking a digestive enzyme supplement containing Xylanase 10 minutes before eating to greatly reduce symptoms.
Phenol sulfur transferase
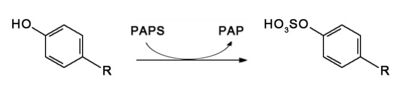
Phenol sulfur transferase, in short PST or SULT1, is a subfamily of the enzyme cytosolic sulfotransferases (SULTs) consisting of at least 8 isoforms in humans[1] that catalyze the transfer of sulfuryl group from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to phenolic compounds,[2] resulting in more hydrophilic products that can be more easily expelled from tissues for excretion.[3] At high concentration, PST could also catalyze the sulfate conjugation of amino groups.[4] This enzyme subfamily, which exists in nearly all human tissues,[5] is important for the detoxification of phenol-containing xenobiotics or endogenous compounds,[6] including the biotransformation of neurotransmitters and drugs.[5] Its expression is controlled by the PST genes located on chromosomes 2, 4, and 16 depending on the isoform,[7] for example the genes for the predominant isoform throughout the body of human adults, SULT1A1,[8][9] which is highly heritable and variable between individuals,[10] and the most important one in the nervous system, SULT1A3,[11] are located on chromosome 16 at the position of 16p11.2 to 16p12.1.[12]
Discovery
PST deficiency was first discovered in the late 1990s by Dr. Rosemary Waring through a series of tests during her researches on the mechanisms and characteristics of sulfation in autistic children.[13] From the result of the test administering individuals with paracetamol, it was found that the level of sulfate conjugate in urine was significantly lower in the autistic individuals as compared to the non-autistic controls, which was caused by the decreased ability in the formation of sulfated metabolites.[14] The level of sulfate in plasma was also found to be significantly lower in autistic children, leading to a reduced activity of PST.[15] Therefore, she concluded that there was possibly a deficiency of PST in autistic children due to the reduction of sulfate in plasma as a substrate of PST.[13]
Pathophysiology
Causes
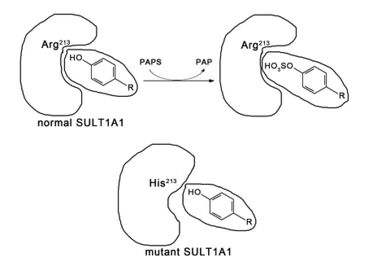
PST deficiency can be caused by inherited mutations in the PST genes,[10] for example the SULT1A1*2 polymorphism, which is a single nucleotide polymorphism at the 638th base of the SULT1A1 gene from guanine to adenosine that causes the change of the 213th amino acid residue of the resultant SULT1A1 from arginine to histidine.[17][18] This mutation causes a conformational change in the enzyme, reducing the size of the binding site and altering the thermochemical properties, which halves the substrate binding affinity and enzyme thermostability,[19] and results in diminished enzymatic activity.[16]
The methylation at the distal and proximal promoters of the PST gens is another mutation that accounts for the deficiency, which causes a reduction in PST expression rather than conformational abnormalities. This prevents the binding of RNA polymerase, which therefore inhibits the mRNA expression of the gene for the production of PST, and finally results in PST deficiency.[20]
Disease-causing mechanisms
PST deficiency can directly cause diseases by the resulted phenol sulfoconjugation defect which reduces the removal of toxic phenolic compounds.[21]
In the liver, where PST serves as one of the important enzymes involved in detoxification, the reduced transcriptional and translational levels of the PST genes would lead to the accumulation of phenolic xenobiotics and cause liver diseases like hepatic steatosis and cirrhosis,[22] or even liver cancers like hepatocellular carcinoma when phenolic carcinogens are accumulated to trigger their developments.[23]
In clinical neurochemistry, PST, in particular the SULT1A3 isoform, is responsible for the degradation of phenolic neurotransmitters such as dopamine and norepinephrine, and therefore is important in the regulation of neurotransmitters which would greatly affect neurological functions. Deficiency or down-regulation of SULT1A3 would cause the retention of neurotransmitter in synapses which affects brain functions including cognitive flexibility and associative learning.[11][24]
Clinical impact
Related disorders
Symptoms of PST deficiency are mainly resulted from the disruptions in multiple metabolic processes due to the accumulation of phenols in the body. Common symptoms include polydipsia, flushing, tachycardia, night sweats, and gastrointestinal problems such as diarrhoea.[13] Neurological and psychiatric disorders such as depression may also occur when regulation of phenolic neurotransmitters is disrupted.[25] PST deficiency is also a risk factor for various diseases including autism, migraine, and cancers.
Autism
It is suspected that mutations, including both microdeletion and microduplication, of the PST genes are the risk factors of autism spectrum disorder,[26] especially the mutation causing decreased SULT1A activity which is usually reported in autistic individuals.[1] Some studies have found that sulfotransferases like PST are involved in glycosylation, and therefore PST deficiency may cause impaired glycosylation, leading to dystroglycanopathies where severe abnormalities of the central nervous system including neuronal migration and cortical defects would occur, and finally result in autistic behaviours.[27] However, it is still unclear on whether PST deficiency is a cause of autism, or just a biomarker for the disorder.[28] Although recent researches have associated autism with the mutations in the position 16p11.2 on chromosome 16,[29][30] where the gene of the predominant PST isoform in the nervous system SULT1A3 exists,[11] due to the large number of gene in this region, PST deficiency resulted from the mutation there may not be a cause of autism but just a condition that is associated with the mutation of another gene which is causing autism.[31][28]
Migraine
PST deficiency in platelets is a risk factor of migraine.[32] It is believed that the reduced PST levels and activity raise the amount of unconjugated amines in the bloodstream and the central nervous system, resulting in a rise of catecholamine level which contributes to the occurrence of recurring headache in migraine.[33] It is also found that dietary intake of foods that are rich in amines may further lower the activity of PST and trigger more serious migraine symptoms.[33]
Cancers
It is controversial for whether PST deficiency increases or decreases the risk of cancers.[34] Although one major function of PST is to inactivate phenolic carcinogens, and therefore a deficiency of PST would reduce inactivation of carcinogens and result in a higher risk of cancer, some studies have also found that PST, specifically SULT1A1, is responsible for the toxification of dietary and environmental mutagens which would increase the risk of cancer, and therefore a decreased risk may be associated with the deficient state of SULT1A1.[19]
Pharmacological impacts
Drug metabolism of phenolic drugs, such as paracetamol and salicylamide, is greatly dependent on the phenol sulfoconjugation by PST, and therefore careful controls on the dosage forms, routes, rates, and duration of administration of those drugs are important for PST deficient patients to prevent accumulation of drugs in the body and depletion of PST for the sulfoconjugation of other xenobiotics and endogenous substances.[35] High dosage of nonsteroidal anti-inflammatory drugs (NSAIDs), such as aspirin, would also cause a short term inhibition to the activity of PST, and should be administered to PST deficient patients with caution to prevent further reduction in PST activity and accumulation of phenolic compounds which would result in adverse impacts.[14]
References
- ↑ 1.0 1.1 "Expression and localization of cytosolic sulfotransferase (SULT) 1A1 and SULT1A3 in normal human brain". Drug Metabolism and Disposition 37 (4): 706–9. April 2009. doi:10.1124/dmd.108.025767. PMID 19171676.
- ↑ "Enzymatic aspects of the phenol (aryl) sulfotransferases". Drug Metabolism Reviews 33 (3–4): 369–95. 2001. doi:10.1081/dmr-120001394. PMID 11768773.
- ↑ "Cloning of the human phenol sulfotransferase gene family: three genes implicated in the metabolism of catecholamines, thyroid hormones and drugs". Chemico-Biological Interactions 109 (1–3): 29–41. February 1998. doi:10.1016/S0009-2797(97)00118-X. PMID 9566731.
- ↑ "Sulphation catalysed by the human cytosolic sulphotransferases--chemical defence or molecular terrorism?". Human & Experimental Toxicology 15 (7): 547–55. July 1996. doi:10.1177/096032719601500701. PMID 8818707.
- ↑ 5.0 5.1 "Phenol sulfotransferase inheritance". Cellular and Molecular Neurobiology 8 (1): 27–34. March 1988. doi:10.1007/bf00712908. PMID 3042142.
- ↑ "Phenol sulfotransferases". The Journal of Biological Chemistry 254 (13): 5658–63. July 1979. doi:10.1016/S0021-9258(18)50465-8. PMID 447677.
- ↑ "Human sulfotransferases and their role in chemical metabolism". Toxicological Sciences 90 (1): 5–22. March 2006. doi:10.1093/toxsci/kfj061. PMID 16322073. https://academic.oup.com/toxsci/article/90/1/5/1692243.
- ↑ "Arginine residues in the active site of human phenol sulfotransferase (SULT1A1)". The Journal of Biological Chemistry 278 (38): 36358–64. September 2003. doi:10.1074/jbc.M306045200. PMID 12867416.
- ↑ "The structure of the catechin-binding site of human sulfotransferase 1A1". Proceedings of the National Academy of Sciences of the United States of America 113 (50): 14312–14317. December 2016. doi:10.1073/pnas.1613913113. PMID 27911811.
- ↑ 10.0 10.1 "Functional genetic variants in the 3'-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity". Toxicological Sciences 118 (2): 391–403. December 2010. doi:10.1093/toxsci/kfq296. PMID 20881232.
- ↑ 11.0 11.1 11.2 "Allosteres to regulate neurotransmitter sulfonation". The Journal of Biological Chemistry 294 (7): 2293–2301. February 2019. doi:10.1074/jbc.RA118.006511. PMID 30545938.
- ↑ "Mapping of the phenol sulfotransferase gene (STP) to human chromosome 16p12.1-p11.2 and to mouse chromosome 7". Genomics 18 (2): 440–3. November 1993. doi:10.1006/geno.1993.1494. PMID 8288252.
- ↑ 13.0 13.1 13.2 "Enzyme and sulphur oxidation deficiencies in autistic children with known food intolerances". Journal of Orthomolecular Medicine 8 (4): 198–200. 1993. http://orthomolecular.org/library/jom/1993/pdf/1993-v08n04-p198.pdf.
- ↑ 14.0 14.1 "Sulphation deficit in "low-functioning" autistic children: a pilot study". Biological Psychiatry 46 (3): 420–4. August 1999. doi:10.1016/s0006-3223(98)00337-0. PMID 10435209.
- ↑ "Biochemical parameters in autistic children". Dev Brain Dysfunct 10: 43–47. 1997. ISSN 1019-5815. https://www.scopus.com/record/display.uri?eid=2-s2.0-0031052835&origin=inward&txGid=e3896cf9d9638ea4bc2a5c112fcbbfe1.
- ↑ 16.0 16.1 "Structural and Dynamic Characterizations Highlight the Deleterious Role of SULT1A1 R213H Polymorphism in Substrate Binding". International Journal of Molecular Sciences 20 (24): 6256. December 2019. doi:10.3390/ijms20246256. PMID 31835852.
- ↑ "Cytochrome and sulfotransferase gene variation in north African populations". Pharmacogenomics 17 (13): 1415–23. August 2016. doi:10.2217/pgs-2016-0016. PMID 27471773.
- ↑ "A functional polymorphism in the SULT1A1 gene (G638A) is associated with risk of lung cancer in relation to tobacco smoking". Carcinogenesis 25 (5): 773–8. May 2004. doi:10.1093/carcin/bgh053. PMID 14688021.
- ↑ 19.0 19.1 "Case-control study and meta-analysis of SULT1A1 Arg213His polymorphism for gene, ethnicity and environment interaction for cancer risk". British Journal of Cancer 99 (8): 1340–7. October 2008. doi:10.1038/sj.bjc.6604683. PMID 18854828.
- ↑ "Epigenetic silencing of the sulfotransferase 1A1 gene by hypermethylation in breast tissue". Oncology Reports 15 (1): 27–32. January 2006. doi:10.3892/or.15.1.27. PMID 16328031.
- ↑ "Sulfoconjugation of catecholamines, nutrition, and hypertension". Hypertension 4 (5 Pt 2): III93-8. 1982. doi:10.1161/01.HYP.4.5_Pt_2.III93. PMID 7049935.
- ↑ "Downregulation of sulfotransferase expression and activity in diseased human livers". Drug Metabolism and Disposition 41 (9): 1642–50. September 2013. doi:10.1124/dmd.113.050930. PMID 23775849.
- ↑ "The loss of phenol sulfotransferase 1 in hepatocellular carcinogenesis". Proteomics 10 (2): 266–76. January 2010. doi:10.1002/pmic.200900721. PMID 19904771.
- ↑ "Sources and physiological significance of plasma dopamine sulfate". The Journal of Clinical Endocrinology and Metabolism 84 (7): 2523–31. July 1999. doi:10.1210/jcem.84.7.5864. PMID 10404831.
- ↑ "Assay of phenol sulphotransferase in human blood". Clinica Chimica Acta; International Journal of Clinical Chemistry 134 (1–2): 199–206. October 1983. doi:10.1016/0009-8981(83)90197-3. PMID 6580977.
- ↑ "Association between microdeletion and microduplication at 16p11.2 and autism". The New England Journal of Medicine 358 (7): 667–75. February 2008. doi:10.1056/NEJMoa075974. PMID 18184952.
- ↑ "Glycan susceptibility factors in autism spectrum disorders". Molecular Aspects of Medicine 51: 104–14. October 2016. doi:10.1016/j.mam.2016.07.001. PMID 27418189.
- ↑ 28.0 28.1 "Diagnostic and Severity-Tracking Biomarkers for Autism Spectrum Disorder". Journal of Molecular Neuroscience 66 (4): 492–511. December 2018. doi:10.1007/s12031-018-1192-1. PMID 30357679.
- ↑ "16p11.2 deletion and duplication: Characterizing neurologic phenotypes in a large clinically ascertained cohort". American Journal of Medical Genetics. Part A 170 (11): 2943–2955. November 2016. doi:10.1002/ajmg.a.37820. PMID 27410714.
- ↑ "Hyperactivity and male-specific sleep deficits in the 16p11.2 deletion mouse model of autism". Autism Research 10 (4): 572–584. April 2017. doi:10.1002/aur.1707. PMID 27739237.
- ↑ "Reciprocal Effects on Neurocognitive and Metabolic Phenotypes in Mouse Models of 16p11.2 Deletion and Duplication Syndromes". PLOS Genetics 12 (2): e1005709. February 2016. doi:10.1371/journal.pgen.1005709. PMID 26872257.
- ↑ "Platelet phenolsulphotransferase deficiency in dietary migraine". Lancet 1 (8279): 983–6. May 1982. doi:10.1016/S0140-6736(82)91990-0. PMID 6122845.
- ↑ 33.0 33.1 "Platelet sulphotransferase activity, plasma sulphate levels and sulphation capacity in patients with migraine and tension headache". Cephalalgia 17 (7): 761–4. November 1997. doi:10.1046/j.1468-2982.1997.1707761.x. PMID 9399006.
- ↑ "Relationship of SULT1A1 copy number variation with estrogen metabolism and human health". The Journal of Steroid Biochemistry and Molecular Biology 174: 169–175. November 2017. doi:10.1016/j.jsbmb.2017.08.017. PMID 28867356.
- ↑ "Sulfate conjugation in drug metabolism: role of inorganic sulfate". Federation Proceedings 45 (8): 2235–40. July 1986. PMID 3459670.
 |