Chemistry:Tracheal cytotoxin
| |
| Names | |
|---|---|
| IUPAC name
(2R,6S)-6-[[(4R)-4-[[(2S)-2-[[(2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoyl]amino]propanoyl]amino]-4-carboxybutanoyl]amino]-2-amino-7-[[(1R)-1-carboxyethyl]amino]-7-oxoheptanoic acid
| |
Other names
| |
| Identifiers | |
| |
3D model (JSmol)
|
|
| DrugBank | |
PubChem CID
|
|
| |
| Properties | |
| C37H59N7O20 | |
| Molar mass | 921.908 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
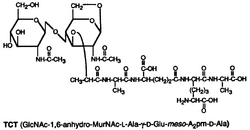
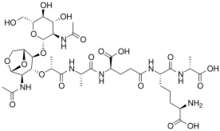
Tracheal cytotoxin (TCT) is a 921 dalton glycopeptide released by Bordetella pertussis,[1] Vibrio fischeri (as a symbiosis chemical),[2] and Neisseria gonorrhoeae (among other peptidoglycan-derived cytotoxins it produces).[3] It is a soluble piece of peptidoglycan (PGN) found in the cell wall of all gram-negative bacteria,[4] but only some bacteria species release TCT due to inability to recycle this piece of anhydromuropeptide.[5]
History
In 1980, it was discovered that B. pertussis could attach to hamster tracheal epithelial (HTE) cells, and also, that the supernatant from the cultured bacterium could disrupt the cell cycle of uninfected cells.[6] This prompted the scientists W. E. Goldman, D. G. Klapper, and J. B. Baseman to isolate and characterize a novel substance from B. pertussis supernatant. The novel disaccharide tetrapeptide that they had purified showed toxicity for HTE cells and tracheal ring cultures. Subsequently, they named the newly sequestered molecule tracheal cytotoxin (TCT).[7]
Structure
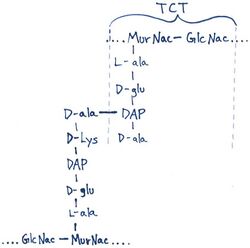
TCT is a soluble piece of peptidoglycan (PGN) found in the cell wall of all gram-negative bacteria.[4] Like all PGNs, TCT is composed of a disaccharide and a peptide chain. The IUPAC name for TCT is N-acetylglucosaminyl-1,6-anhydro-N-acetylmuramyl-(L)-alanyl-γ-(D)-glutamyl-mesodiaminopimelyl-(D)-alanine.[8] It is classified as a DAP (diaminopimelic acid)-type PGN due to the third amino group within the chain being a diaminopimelyl peptide.[citation needed]
The DAP residue is responsible for directly bonding to the D-alanine peptide of another PGN molecule, thus aiding TCT's attachment within the cell wall.[9]
The DAP portion of TCT also implies importance in cytopathogenicity as analogs lacking DAP show a significant reduction in toxicity.[10]
Mechanism of pathogenesis
Most Gram-negative bacteria keep TCT within the cell wall by using a PGN-transporter protein known as AmpG. However, B. pertussis is not capable of recycling PGNs via AmpG and thus, TCT escapes into the surrounding environment.[11][5] Also, TCT appears to be constitutively expressed[dubious ] by B. pertussis.[4]
The first murine-model studies using TCT involved treatment of hamster tracheal cells. These experiments alluded to TCT's role in ciliostasis and cellular extrusion of ciliated hamster cells. Also, HTE cells had a markedly reduced level of DNA synthesis post-treatment with TCT.
While previous studies using murine models reported evidence of TCT causing ciliostasis, in vitro studies using human tracheal cells have shown that TCT does not affect ciliary beat frequency of living cells, but instead causes damage and eventual extrusion of ciliated cells.[12] In gonorrhea infections, vaginal ciliated epithelial cells have also displayed the same cytopathogenic effects due to TCT recognition.[13] The extensive damage to ciliated epithelial tissue caused by TCT results in major disruption to the ciliary escalator; an important asset of the host's non-specific defenses. This disruption hinders the host's ability to remove mucous and foreign microbes from the epithelial tissue. Paroxysmal cough, e.g. whooping cough, is a direct symptom of said mucous build-up due to ciliated tissue damage.[citation needed]
NOD-1 recognition and the presence of Lipooligosaccharide (LOS) are two factors that modulate the effect of TCT. NOD-1 is a pattern recognition receptor that detects peptidoglycan. This receptor reacts weakly to TCT in humans, but robustly in mice. TCT is thought to work synergistically with LOS to mediate an inflammatory response, thus causing damage to ciliated epithelial cells.[14] Notably, the human pathogens (B. pertussis and N. gonorrhea) that produce excess TCT, causing damage to cilia also both produce LOS in their outer membrane.[citation needed]
Effect on immune system
TCT has been classified as an adjuvant molecule because of the stimulating effects it has on the immune system. Cellular damage associated with TCT is thought to be a result of increased levels of nitric oxide (NO) secretion by mucosal cells as part of an innate defense response to extracellular lipopolysaccharide (LPS) and TCT.[15] In humans, peptidoglycan recognition proteins, e.g. PGRPIαC, appear to bind with TCT and consequently induce the Tumor Necrosis Factor Receptor (TNFR) pathway.[16] Studies using murine macrophages have shown that TCT encourages cytokine secretion, probably through the NOD1 receptor.[17] As a pleiotropic toxin, TCT also acts as a pyrogen and as a stimulant of slow-wave sleep.[18]
Peptidoglycan recognition protein 4 (PGLYRP4), in mammals (mice), interacts with TCT and reduces damage from pertussis inflammation.[19] This molecule has similar immune-eliciting properties in Drosophila, where a pair of PGRPs perform the recognition.[20]
References
- ↑ Cundell, DR; Kanthakumar, K; Taylor, GW; Goldman, WE; Flak, T; Cole, PJ; Wilson, R (1994). "Effect of tracheal cytotoxin from Bordetella pertussis on human neutrophil function in vitro". Infection and Immunity 62 (2): 639–43. doi:10.1128/iai.62.2.639-643.1994. PMID 8300220.
- ↑ Koropatnick, TA; Engle, JT; Apicella, MA; Stabb, EV; Goldman, WE; McFall-Ngai, MJ (12 November 2004). "Microbial factor-mediated development in a host-bacterial mutualism.". Science 306 (5699): 1186–8. doi:10.1126/science.1102218. PMID 15539604. Bibcode: 2004Sci...306.1186K. "Here, we report that Vibrio fischeri also releases TCT, which acts in synergy with lipopolysaccharide (LPS) to trigger tissue development in its mutualistic symbiosis with the squid Euprymna scolopes.".
- ↑ Cloud, KA; Dillard, JP (June 2002). "A lytic transglycosylase of Neisseria gonorrhoeae is involved in peptidoglycan-derived cytotoxin production.". Infection and Immunity 70 (6): 2752–7. doi:10.1128/IAI.70.6.2752-2757.2002. PMID 12010959. "A major peptidoglycan fragment released by gonococci is identical to the tracheal cytotoxin of Bordetella pertussis and has been shown to kill ciliated fallopian tube cells in organ culture.".
- ↑ 4.0 4.1 4.2 Mattoo, S.; Cherry, J. D. (2005). "Molecular Pathogenesis, Epidemiology, and Clinical Manifestations of Respiratory Infections Due to Bordetella pertussis and Other Bordetella Subspecies". Clinical Microbiology Reviews 18 (2): 326–82. doi:10.1128/CMR.18.2.326-382.2005. PMID 15831828.
- ↑ 5.0 5.1 Parkhill, Julian; Sebaihia, Mohammed; Preston, Andrew; Murphy, Lee D; Thomson, Nicholas; Harris, David E; Holden, Matthew T G; Churcher, Carol M et al. (2003). "Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica". Nature Genetics 35 (1): 32–40. doi:10.1038/ng1227. PMID 12910271.
- ↑ Goldman, W.E. and J.B. Baseman (March 1980). "Abstr. Ann. Meet. Am. Soc. Microbiol.". Abstr. Ann. Meet. Am. Soc. Microbiol..[page needed]
- ↑ Goldman, William E.; Klapper, David G.; Baseman, Joel B. (1982-05-01). "Detection, isolation, and analysis of a released Bordetella pertussis product toxic to cultured tracheal cells". Infection and Immunity 36 (2): 782–94. doi:10.1128/iai.36.2.782-794.1982. PMID 6177637.
- ↑ Cookson, BT; Tyler, AN; Goldman, WE (1989). "Primary structure of the peptidoglycan-derived tracheal cytotoxin of Bordetella pertussis". Biochemistry 28 (4): 1744–9. doi:10.1021/bi00430a048. PMID 2541765.
- ↑ Wilson, R; Read, R; Thomas, M; Rutman, A; Harrison, K; Lund, V; Cookson, B; Goldman, W et al. (1991-01-01). "Effects of Bordetella pertussis infection on human respiratory epithelium in vivo and in vitro". Infection and Immunity 59 (1): 337–45. doi:10.1128/iai.59.1.337-345.1991. PMID 1987048.
- ↑ Luker, K. E.; Collier, J. L.; Kolodziej, E. W.; Marshall, G. R.; Goldman, W. E. (1993). "Bordetella pertussis Tracheal Cytotoxin and Other Muramyl Peptides: Distinct Structure-Activity Relationships for Respiratory Epithelial Cytopathology". Proceedings of the National Academy of Sciences 90 (6): 2365–2369. doi:10.1073/pnas.90.6.2365. PMID 8460147. Bibcode: 1993PNAS...90.2365L.
- ↑ Jacobs, C; Joris, B; Jamin, M; Klarsov, K; Van Beeumen, J; Mengin-Lecreulx, D; Van Heijenoort, J; Park, JT et al. (1995). "AmpD, essential for both beta-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-L-alanine amidase". Molecular Microbiology 15 (3): 553–9. doi:10.1111/j.1365-2958.1995.tb02268.x. PMID 7783625.
- ↑ Wilson, R; Read, R; Thomas, M; Rutman, A; Harrison, K; Lund, V; Cookson, B; Goldman, W et al. (1991). "Effects of Bordetella pertussis infection on human respiratory epithelium in vivo and in vitro". Infection and Immunity 59 (1): 337–45. doi:10.1128/iai.59.1.337-345.1991. PMID 1987048.
- ↑ Swaminathan, C. P.; Brown, P. H.; Roychowdhury, A.; Wang, Q.; Guan, R.; Silverman, N.; Goldman, W. E.; Boons, G.-J. et al. (2006). "Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs)". Proceedings of the National Academy of Sciences 103 (3): 684–9. doi:10.1073/pnas.0507656103. PMID 16407132. Bibcode: 2006PNAS..103..684S.
- ↑ Melvin, Jeffrey A.; Scheller, Erich V.; Miller, Jeff F.; Cotter, Peggy A. (2014-04-01). "Bordetella pertussis pathogenesis: current and future challenges" (in en). Nature Reviews Microbiology 12 (4): 274–288. doi:10.1038/nrmicro3235. ISSN 1740-1526. PMID 24608338.
- ↑ Coote, JG (2001). Environmental sensing mechanisms in Bordetella. Advances in Microbial Physiology. 44. pp. 141–81. doi:10.1016/S0065-2911(01)44013-6. ISBN 9780120277445.
- ↑ Swaminathan, C. P.; Brown, PH; Roychowdhury, A; Wang, Q; Guan, R; Silverman, N; Goldman, WE; Boons, GJ et al. (2006). "Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs)". Proceedings of the National Academy of Sciences 103 (3): 684–9. doi:10.1073/pnas.0507656103. PMID 16407132. Bibcode: 2006PNAS..103..684S.
- ↑ Humann, Jessica; Lenz, Laurel L. (2009). "Bacterial Peptidoglycan-Degrading Enzymes and Their Impact on Host Muropeptide Detection". Journal of Innate Immunity 1 (2): 88–97. doi:10.1159/000181181. PMID 19319201.
- ↑ Magalhaes, Joao Gamelas; Philpott, Dana J; Nahori, Marie-Anne; Jéhanno, Muguette; Fritz, Joerg; Bourhis, Lionel Le; Viala, Jérôme; Hugot, Jean-Pierre et al. (2005). "Murine Nod1 but not its human orthologue mediates innate immune detection of tracheal cytotoxin". EMBO Reports 6 (12): 1201–7. doi:10.1038/sj.embor.7400552. PMID 16211083.
- ↑ Skerry, C; Goldman, WE; Carbonetti, NH (February 2019). "Peptidoglycan Recognition Protein 4 Suppresses Early Inflammatory Responses to Bordetella pertussis and Contributes to Sphingosine-1-Phosphate Receptor Agonist-Mediated Disease Attenuation.". Infection and Immunity 87 (2). doi:10.1128/IAI.00601-18. PMID 30510103.
- ↑ Chang, CI; Chelliah, Y; Borek, D; Mengin-Lecreulx, D; Deisenhofer, J (24 March 2006). "Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor.". Science 311 (5768): 1761–4. doi:10.1126/science.1123056. PMID 16556841. Bibcode: 2006Sci...311.1761C.
External links
- Ligand entry in the Protein Data Bank: MLD
- Entry in MetaCyc: CPD0-1080
 |