Manifold regularization
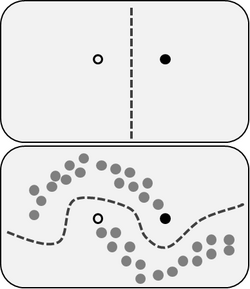
In machine learning, manifold regularization is a technique for using the shape of a dataset to constrain the functions that should be learned on that dataset. In many machine learning problems, the data to be learned do not cover the entire input space. For example, a facial recognition system may not need to classify any possible image, but only the subset of images that contain faces. The technique of manifold learning assumes that the relevant subset of data comes from a manifold, a mathematical structure with useful properties. The technique also assumes that the function to be learned is smooth: data with different labels are not likely to be close together, and so the labeling function should not change quickly in areas where there are likely to be many data points. Because of this assumption, a manifold regularization algorithm can use unlabeled data to inform where the learned function is allowed to change quickly and where it is not, using an extension of the technique of Tikhonov regularization. Manifold regularization algorithms can extend supervised learning algorithms in semi-supervised learning and transductive learning settings, where unlabeled data are available. The technique has been used for applications including medical imaging, geographical imaging, and object recognition.
Manifold regularizer
Motivation
Manifold regularization is a type of regularization, a family of techniques that reduces overfitting and ensures that a problem is well-posed by penalizing complex solutions. In particular, manifold regularization extends the technique of Tikhonov regularization as applied to Reproducing kernel Hilbert spaces (RKHSs). Under standard Tikhonov regularization on RKHSs, a learning algorithm attempts to learn a function from among a hypothesis space of functions . The hypothesis space is an RKHS, meaning that it is associated with a kernel , and so every candidate function has a norm , which represents the complexity of the candidate function in the hypothesis space. When the algorithm considers a candidate function, it takes its norm into account in order to penalize complex functions.
Formally, given a set of labeled training data with and a loss function , a learning algorithm using Tikhonov regularization will attempt to solve the expression
where is a hyperparameter that controls how much the algorithm will prefer simpler functions over functions that fit the data better.

Manifold regularization adds a second regularization term, the intrinsic regularizer, to the ambient regularizer used in standard Tikhonov regularization. Under the manifold assumption in machine learning, the data in question do not come from the entire input space , but instead from a nonlinear manifold . The geometry of this manifold, the intrinsic space, is used to determine the regularization norm.[1]
Laplacian norm
There are many possible choices for the intrinsic regularizer . Many natural choices involve the gradient on the manifold , which can provide a measure of how smooth a target function is. A smooth function should change slowly where the input data are dense; that is, the gradient should be small where the marginal probability density , the probability density of a randomly drawn data point appearing at , is large. This gives one appropriate choice for the intrinsic regularizer:
In practice, this norm cannot be computed directly because the marginal distribution is unknown, but it can be estimated from the provided data.
Graph-based approach of the Laplacian norm
When the distances between input points are interpreted as a graph, then the Laplacian matrix of the graph can help to estimate the marginal distribution. Suppose that the input data include labeled examples (pairs of an input and a label ) and unlabeled examples (inputs without associated labels). Define to be a matrix of edge weights for a graph, where is a similarity built from distance measure between the data points and (so that more close implies higher ). Define to be a diagonal matrix with and to be the Laplacian matrix . Then, as the number of data points increases, converges to the Laplace–Beltrami operator , which is the divergence of the gradient .[2][3] Then, if is a vector of the values of at the data, , the intrinsic norm can be estimated:
As the number of data points increases, this empirical definition of converges to the definition when is known.[1]
Solving the regularization problem with graph-based approach
Using the weights and for the ambient and intrinsic regularizers, the final expression to be solved becomes:
As with other kernel methods, may be an infinite-dimensional space, so if the regularization expression cannot be solved explicitly, it is impossible to search the entire space for a solution. Instead, a representer theorem shows that under certain conditions on the choice of the norm , the optimal solution must be a linear combination of the kernel centered at each of the input points: for some weights ,
Using this result, it is possible to search for the optimal solution by searching the finite-dimensional space defined by the possible choices of .[1]
Functional approach of the Laplacian norm
The idea beyond the graph-Laplacian is to use neighbors to estimate the Laplacian. This method is akin to local averaging methods, that are known to scale poorly in high-dimensional problems. Indeed, the graph Laplacian is known to suffer from the curse of dimensionality.[2] Luckily, it is possible to leverage expected smoothness of the function to estimate thanks to more advanced functional analysis. This method consists of estimating the Laplacian operator using derivatives of the kernel reading where denotes the partial derivatives according to the j-th coordinate of the first variable.[4] This second approach to the Laplacian norm is to put in relation with meshfree methods, that contrast with the finite difference method in PDE.
Applications
Manifold regularization can extend a variety of algorithms that can be expressed using Tikhonov regularization, by choosing an appropriate loss function and hypothesis space . Two commonly used examples are the families of support vector machines and regularized least squares algorithms. (Regularized least squares includes the ridge regression algorithm; the related algorithms of LASSO and elastic net regularization can be expressed as support vector machines.[5][6]) The extended versions of these algorithms are called Laplacian Regularized Least Squares (abbreviated LapRLS) and Laplacian Support Vector Machines (LapSVM), respectively.[1]
Laplacian Regularized Least Squares (LapRLS)
Regularized least squares (RLS) is a family of regression algorithms: algorithms that predict a value for its inputs , with the goal that the predicted values should be close to the true labels for the data. In particular, RLS is designed to minimize the mean squared error between the predicted values and the true labels, subject to regularization. Ridge regression is one form of RLS; in general, RLS is the same as ridge regression combined with the kernel method. The problem statement for RLS results from choosing the loss function in Tikhonov regularization to be the mean squared error:
Thanks to the representer theorem, the solution can be written as a weighted sum of the kernel evaluated at the data points:
and solving for gives:
where is defined to be the kernel matrix, with , and is the vector of data labels.
Adding a Laplacian term for manifold regularization gives the Laplacian RLS statement:
The representer theorem for manifold regularization again gives
and this yields an expression for the vector . Letting be the kernel matrix as above, be the vector of data labels, and be the block matrix :
with a solution of
LapRLS has been applied to problems including sensor networks,[7] medical imaging,[8][9] object detection,[10] spectroscopy,[11] document classification,[12] drug-protein interactions,[13] and compressing images and videos.[14]
Laplacian Support Vector Machines (LapSVM)
Support vector machines (SVMs) are a family of algorithms often used for classifying data into two or more groups, or classes. Intuitively, an SVM draws a boundary between classes so that the closest labeled examples to the boundary are as far away as possible. This can be directly expressed as a linear program, but it is also equivalent to Tikhonov regularization with the hinge loss function, :
Adding the intrinsic regularization term to this expression gives the LapSVM problem statement:
Again, the representer theorem allows the solution to be expressed in terms of the kernel evaluated at the data points:
can be found by writing the problem as a linear program and solving the dual problem. Again letting be the kernel matrix and be the block matrix , the solution can be shown to be
where is the solution to the dual problem
and is defined by
LapSVM has been applied to problems including geographical imaging,[17][18][19] medical imaging,[20][21][22] face recognition,[23] machine maintenance,[24] and brain–computer interfaces.[25]
Limitations
- Manifold regularization assumes that data with different labels are not likely to be close together. This assumption is what allows the technique to draw information from unlabeled data, but it only applies to some problem domains. Depending on the structure of the data, it may be necessary to use a different semi-supervised or transductive learning algorithm.[26]
- In some datasets, the intrinsic norm of a function can be very close to the ambient norm : for example, if the data consist of two classes that lie on perpendicular lines, the intrinsic norm will be equal to the ambient norm. In this case, unlabeled data have no effect on the solution learned by manifold regularization, even if the data fit the algorithm's assumption that the separator should be smooth. Approaches related to co-training have been proposed to address this limitation.[27]
- If there are a very large number of unlabeled examples, the kernel matrix becomes very large, and a manifold regularization algorithm may become prohibitively slow to compute. Online algorithms and sparse approximations of the manifold may help in this case.[28]
See also
- Manifold learning
- Manifold hypothesis
- Semi-supervised learning
- Transduction (machine learning)
- Spectral graph theory
- Reproducing kernel Hilbert space
- Tikhonov regularization
- Differential geometry
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Belkin, Mikhail; Niyogi, Partha; Sindhwani, Vikas (2006). "Manifold regularization: A geometric framework for learning from labeled and unlabeled examples". The Journal of Machine Learning Research 7: 2399–2434. http://dl.acm.org/citation.cfm?id=1248632. Retrieved 2015-12-02.
- ↑ 2.0 2.1 Hein, Matthias; Audibert, Jean-Yves; Von Luxburg (2005). "From graphs to manifolds–weak and strong pointwise consistency of graph laplacians". Learning theory. Lecture Notes in Computer Science. 3559. Springer. pp. 470–485. doi:10.1007/11503415_32. ISBN 978-3-540-26556-6.
- ↑ Belkin, Mikhail; Niyogi, Partha (2005). "Towards a theoretical foundation for Laplacian-based manifold methods". Learning theory. Lecture Notes in Computer Science. 3559. Springer. pp. 486–500. doi:10.1007/11503415_33. ISBN 978-3-540-26556-6.
- ↑ Cabannes, Vivien; Pillaud-Vivien, Loucas; Bach, Francis; Rudi, Alessandro (2021). "Overcoming the curse of dimensionality with Laplacian regularization in semi-supervised learning". arXiv:2009.04324 [stat.ML].
- ↑ Jaggi, Martin (2014). An Equivalence between the Lasso and Support Vector Machines. Chapman and Hall/CRC.
- ↑ Zhou, Quan; Chen, Wenlin; Song, Shiji; Gardner, Jacob; Weinberger, Kilian; Chen, Yixin. "A Reduction of the Elastic Net to Support Vector Machines with an Application to GPU Computing". Association for the Advancement of Artificial Intelligence. https://www.aaai.org/ocs/index.php/AAAI/AAAI15/paper/view/9856.
- ↑ Pan, Jeffrey Junfeng; Yang, Qiang; Chang, Hong; Yeung, Dit-Yan (2006). "A manifold regularization approach to calibration reduction for sensor-network based tracking". 21. Menlo Park, CA; Cambridge, MA; London; AAAI Press; MIT Press; 1999. pp. 988. http://www.aaai.org/Papers/AAAI/2006/AAAI06-155.pdf. Retrieved 2015-12-02.
- ↑ Zhang, Daoqiang; Shen, Dinggang (2011). "Semi-supervised multimodal classification of Alzheimer's disease". IEEE. pp. 1628–1631. doi:10.1109/ISBI.2011.5872715.
- ↑ Park, Sang Hyun; Gao, Yaozong; Shi, Yinghuan; Shen, Dinggang (2014). "Interactive Prostate Segmentation Based on Adaptive Feature Selection and Manifold Regularization". Machine Learning in Medical Imaging. Lecture Notes in Computer Science. 8679. Springer. pp. 264–271. doi:10.1007/978-3-319-10581-9_33. ISBN 978-3-319-10580-2.
- ↑ Pillai, Sudeep. Semi-supervised Object Detector Learning from Minimal Labels. http://people.csail.mit.edu/spillai/data/papers/ssl-cv-project-paper.pdf. Retrieved 2015-12-15.
- ↑ Wan, Songjing; Wu, Di; Liu, Kangsheng (2012). "Semi-Supervised Machine Learning Algorithm in Near Infrared Spectral Calibration: A Case Study on Diesel Fuels". Advanced Science Letters 11 (1): 416–419. doi:10.1166/asl.2012.3044.
- ↑ Wang, Ziqiang; Sun, Xia; Zhang, Lijie; Qian, Xu (2013). "Document Classification based on Optimal Laprls". Journal of Software 8 (4): 1011–1018. doi:10.4304/jsw.8.4.1011-1018.
- ↑ Xia, Zheng; Wu, Ling-Yun; Zhou, Xiaobo; Wong, Stephen TC (2010). "Semi-supervised drug-protein interaction prediction from heterogeneous biological spaces". BMC Systems Biology 4 (Suppl 2): –6. doi:10.1186/1752-0509-4-S2-S6. PMID 20840733.
- ↑ Cheng, Li; Vishwanathan, S. V. N. (2007). "Learning to compress images and videos". ACM. pp. 161–168. http://dl.acm.org/citation.cfm?id=1273517. Retrieved 2015-12-16.
- ↑ Lin, Yi; Wahba, Grace; Zhang, Hao; Lee, Yoonkyung (2002). "Statistical properties and adaptive tuning of support vector machines". Machine Learning 48 (1–3): 115–136. doi:10.1023/A:1013951620650.
- ↑ Wahba, Grace; others (1999). "Support vector machines, reproducing kernel Hilbert spaces and the randomized GACV". Advances in Kernel Methods-Support Vector Learning 6: 69–87.
- ↑ Kim, Wonkook; Crawford, Melba M. (2010). "Adaptive classification for hyperspectral image data using manifold regularization kernel machines". IEEE Transactions on Geoscience and Remote Sensing 48 (11): 4110–4121. doi:10.1109/TGRS.2010.2076287. Bibcode: 2010ITGRS..48.4110K.
- ↑ Camps-Valls, Gustavo; Tuia, Devis; Bruzzone, Lorenzo; Atli Benediktsson, Jon (2014). "Advances in hyperspectral image classification: Earth monitoring with statistical learning methods". IEEE Signal Processing Magazine 31 (1): 45–54. doi:10.1109/msp.2013.2279179. Bibcode: 2014ISPM...31...45C.
- ↑ Gómez-Chova, Luis; Camps-Valls, Gustavo; Muñoz-Marí, Jordi; Calpe, Javier (2007). "Semi-supervised cloud screening with Laplacian SVM". IEEE. pp. 1521–1524. doi:10.1109/IGARSS.2007.4423098.
- ↑ Cheng, Bo; Zhang, Daoqiang; Shen, Dinggang (2012). "Domain transfer learning for MCI conversion prediction". Medical Image Computing and Computer-Assisted Intervention–MICCAI 2012. Lecture Notes in Computer Science. 7510. Springer. pp. 82–90. doi:10.1007/978-3-642-33415-3_11. ISBN 978-3-642-33414-6.
- ↑ Jamieson, Andrew R.; Giger, Maryellen L.; Drukker, Karen; Pesce, Lorenzo L. (2010). "Enhancement of breast CADx with unlabeled dataa)". Medical Physics 37 (8): 4155–4172. doi:10.1118/1.3455704. PMID 20879576. Bibcode: 2010MedPh..37.4155J.
- ↑ Wu, Jiang; Diao, Yuan-Bo; Li, Meng-Long; Fang, Ya-Ping; Ma, Dai-Chuan (2009). "A semi-supervised learning based method: Laplacian support vector machine used in diabetes disease diagnosis". Biology:Interdisciplinary Sciences: Computational Life Sciences 1 (2): 151–155. doi:10.1007/s12539-009-0016-2. PMID 20640829.
- ↑ Wang, Ziqiang; Zhou, Zhiqiang; Sun, Xia; Qian, Xu; Sun, Lijun (2012). "Enhanced LapSVM Algorithm for Face Recognition.". International Journal of Advancements in Computing Technology 4 (17). http://search.ebscohost.com/login.aspx?direct=true&profile=ehost&scope=site&authtype=crawler&jrnl=20058039&AN=98908455&h=8QzzRizi2IKxCZ4EHJjzxbGY%2FQazcifd58fcAGEG17GiFk0wZE59DrEge0xfEGhXRqsBaMwuBNyenVSP6sjwsA%3D%3D&crl=c. Retrieved 2015-12-16.
- ↑ Zhao, Xiukuan; Li, Min; Xu, Jinwu; Song, Gangbing (2011). "An effective procedure exploiting unlabeled data to build monitoring system". Expert Systems with Applications 38 (8): 10199–10204. doi:10.1016/j.eswa.2011.02.078.
- ↑ Zhong, Ji-Ying; Lei, Xu; Yao, D. (2009). "Semi-supervised learning based on manifold in BCI". Journal of Electronics Science and Technology of China 7 (1): 22–26. http://www.journal.uestc.edu.cn/archives/2009/1/7/22-2677907.pdf. Retrieved 2015-12-16.
- ↑ Zhu, Xiaojin (2005). Semi-supervised learning literature survey.
- ↑ Sindhwani, Vikas; Rosenberg, David S. (2008). "An RKHS for multi-view learning and manifold co-regularization". ACM. pp. 976–983. http://dl.acm.org/citation.cfm?id=1390279. Retrieved 2015-12-02.
- ↑ Goldberg, Andrew; Li, Ming; Zhu, Xiaojin (2008). "Online Manifold Regularization: A New Learning Setting and Empirical Study". Machine Learning and Knowledge Discovery in Databases. Lecture Notes in Computer Science. 5211. pp. 393–407. doi:10.1007/978-3-540-87479-9_44. ISBN 978-3-540-87478-2.
External links
Software
- The ManifoldLearn library and the Primal LapSVM library implement LapRLS and LapSVM in MATLAB.
- The Dlib library for C++ includes a linear manifold regularization function.
 |
