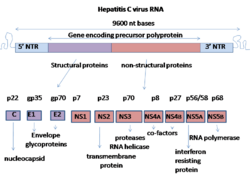
Biology:Hepatitis C virus nonstructural protein 5A
Nonstructural protein 5A (NS5A) is a zinc-binding and proline-rich hydrophilic phosphoprotein that plays a key role in Hepatitis C virus RNA replication.[1][2] It appears to be a dimeric form without trans-membrane helices.[3]
Structure
NS5A is derived from a large polyprotein that is translated from the HCV genome, and undergoes post-translation processing by nonstructural protein 3 (NS3) viral protease.[4] Despite no inherent enzymatic activity being attributed to NS5A, its function is mediated through interaction with other nonstructural (NS) viral and cellular proteins.[2][4] NS5A has two phosphorylated forms: p56 and p58, which differ in the electrophoretic mobility.[3] p56 is basally phosphorylated by host cellular protein kinase at the center and near the C terminus, whereas p58 is a form of hyper-phosphorylated NS5A at the center of the serine-rich region.[3] Protein mass spectrometry identified several phosphorylated serine residues in this region including serine 225, 229, 232, and 235 responsible for NS5A hyper-phosphorylation.[5] An array of phosphorylation-specific antibodies confirmed their phosphorylation in infected cells.[6][7][8][9] It has been predicted that the N-terminal 30 aa of NS5A form an amphipathic α-helix with a highly preserved feature, which is essential to modulate the association between NS5A and ER membrane.[3][4] The IFN-sensitivity determining region (ISDR) at the C-terminal of NS5A has been reported to perform strong trans-activating activities, suggesting that NS5A likely functions as a transcriptional activator.[3]
NS5A has three structurally different domains: Domain I was demonstrated to be an alternative dimeric structure by crystallography, while domain II and III remained unfolded.[1] Furthermore, the conformational flexibility of NS5A plays an important role in multiple HCV infection stages.[1] It is also possible that NS5A is a critical component during HCV replication and subcellular localization, which may shed light on the poorly understood HCV life cycle.[1][4] Additionally, NS5A has been shown to modulate the polymerase activity of NS5B, an RNA-dependent RNA polymerase (RdRp).[3] Intriguingly, NS5A may be a RNA binding protein because it is able to bind to the 3’UTR of the plus and minus HCV RNA strands.[3] Moreover, NS5A is a key mediator in regulating host cell function and activity upon HCV infection.[4] Therefore, NS5A has been extensively studied in HCV research also due to its capability to regulate the interferon (IFN) response of the host cells. Because NS5A exerts functionally essential effects in regulation of viral replication, assembly and egress, it has been considered a potential drug target for antiviral therapeutic intervention.[1][4] Indeed, small molecule drugs efficiently targeting NS5A displayed a much higher potency in controlling HCV infection than other drugs.[1] Therefore, NS5A related researches would have important implications in single molecule drug design and pegIFN-free direct-acting antiviral (DAA) combination therapies.[1]
As a drug target
Many antiviral drugs target NS5A, e.g. to treat hepatitis C; they are often described as NS5A inhibitors, and they are often used in combination with NS5B inhibitors:
- FDA-approved:
- Ledipasvir, approved on October 10, 2014 in a fixed-dose combination (FDC) with sofosbuvir
- Ombitasvir, approved on December 19, 2014 in a FDC with paritaprevir and ritonavir, co-packaged with dasabuvir
- Daclatasvir, approved on July 24, 2015
- Elbasvir, approved on January 28, 2016 in a FDC with grazoprevir
- Velpatasvir, approved on June 28, 2016 in a FDC with sofosbuvir and on July 17, 2017 in a FDC with sofosbuvir and voxilaprevir
- Pibrentasvir, approved on August 3, 2017 in a FDC with glecaprevir
- Investigational drugs:
- Odalasvir
- Ravidasvir
- Ruzasvir
- Samatasvir
Intragenic complementation
Multiple copies of a polypeptide encoded by a gene often can form an aggregate referred to as a multimer. When a multimer is formed from polypeptides produced by two different mutant alleles of a particular gene, the mixed multimer may exhibit greater functional activity than the unmixed multimers formed by each of the mutants alone. When a mixed multimer displays increased functionality relative to the unmixed multimers, the phenomenon is referred to as intragenic complementation.[citation needed]
NS5A protein is a multimer, a dimer in this case, and intragenic complementation of replication-defective NS5A alleles has been demonstrated by Fridell et al.[10] On the bases of pairwise complementation tests between different NS5A mutant alleles, they identified three complementation groups that were considered to define three distinct and genetically separable functions of NS5A in RNA replication.
See also
- Discovery and development of NS5A inhibitors
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Belda, O; Targett-Adams, P (2012). "Small molecule inhibitors of the hepatitis C virus-encoded NS5A protein". Virus Research 170 (1–2): 1–14. doi:10.1016/j.virusres.2012.09.007. PMID 23009750.
- ↑ 2.0 2.1 Huang, Y; Staschke, K; De Francesco, R; Tan, SL (2007). "Phosphorylation of hepatitis C virus NS5A nonstructural protein: A new paradigm for phosphorylation-dependent viral RNA replication?". Virology 364 (1): 1–9. doi:10.1016/j.virol.2007.01.042. PMID 17400273.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 MacDonald, A; Harris, M (2004). "Hepatitis C virus NS5A: Tales of a promiscuous protein". The Journal of General Virology 85 (Pt 9): 2485–502. doi:10.1099/vir.0.80204-0. PMID 15302943.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 He, Y; Staschke, KA; Tan, SL; Tan, SL (2006). "HCV NS5A: A Multifunctional Regulator of Cellular Pathways and Virus Replication". Hepatitis C Viruses: Genomes and Molecular Biology. Horizon Bioscience. https://www.ncbi.nlm.nih.gov/books/NBK1621/. Retrieved 8 January 2021.
- ↑ Chong, Weng Man; Hsu, Shih-Chin; Kao, Wei-Ting; Lo, Chieh-Wen; Lee, Kuan-Ying; Shao, Jheng-Syuan; Chen, Yi-Hung; Chang, Justin et al. (2016-02-19). "Phosphoproteomics Identified an NS5A Phosphorylation Site Involved in Hepatitis C Virus Replication". The Journal of Biological Chemistry 291 (8): 3918–3931. doi:10.1074/jbc.M115.675413. ISSN 1083-351X. PMID 26702051. PMC 4759171. https://pubmed.ncbi.nlm.nih.gov/26702051/.
- ↑ Hsu, Shih-Chin; Lo, Chieh-Wen; Pan, Ting-Chun; Lee, Kuan-Ying; Yu, Ming-Jiun (2017-07-15). "Serine 235 Is the Primary NS5A Hyperphosphorylation Site Responsible for Hepatitis C Virus Replication". Journal of Virology 91 (14). doi:10.1128/JVI.00194-17. ISSN 1098-5514. PMID 28446668. PMC 5487554. https://pubmed.ncbi.nlm.nih.gov/28446668/.
- ↑ Hsu, Shih-Chin; Tsai, Chia-Ni; Lee, Kuan-Ying; Pan, Ting-Chun; Lo, Chieh-Wen; Yu, Ming-Jiun (2018-10-15). "Sequential S232/S235/S238 Phosphorylation of the Hepatitis C Virus Nonstructural Protein 5A". Journal of Virology 92 (20). doi:10.1128/JVI.01295-18. ISSN 1098-5514. PMID 30089697. PMC 6158443. https://pubmed.ncbi.nlm.nih.gov/30089697/.
- ↑ Tsai, Chia-Ni; Pan, Ting-Chun; Chiang, Cho-Han; Yu, Chun-Chiao; Su, Shih-Han; Yu, Ming-Jiun (2019-12-01). "Serine 229 Balances the Hepatitis C Virus Nonstructural Protein NS5A between Hypo- and Hyperphosphorylated States". Journal of Virology 93 (23). doi:10.1128/JVI.01028-19. ISSN 1098-5514. PMID 31511391. PMC 6854479. https://pubmed.ncbi.nlm.nih.gov/31511391/.
- ↑ Chiang, Cho-Han; Lai, Yen-Ling; Huang, Yu-Ning; Yu, Chun-Chiao; Lu, Christine C.; Yu, Guann-Yi; Yu, Ming-Jiun (2020-09-15). "Sequential Phosphorylation of the Hepatitis C Virus NS5A Protein Depends on NS3-Mediated Autocleavage between NS3 and NS4A". Journal of Virology 94 (19). doi:10.1128/JVI.00420-20. ISSN 1098-5514. PMID 32699091. PMC 7495366. https://pubmed.ncbi.nlm.nih.gov/32699091/.
- ↑ Fridell RA, Valera L, Qiu D, Kirk MJ, Wang C, Gao M. Intragenic complementation of hepatitis C virus NS5A RNA replication-defective alleles. J Virol. 2013;87(4):2320-2329. doi:10.1128/JVI.02861-12
 |