Biology:DNA adenine methylase
| Site-specific DNA-methyltransferase (adenine-specific) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.1.1.72 | ||||||||
| CAS number | 69553-52-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
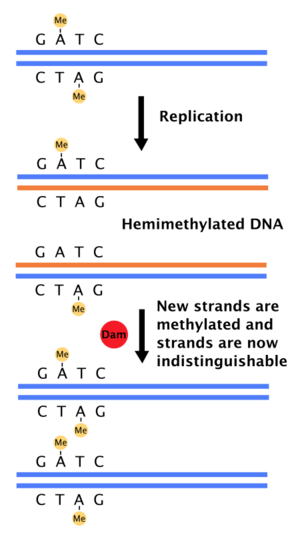
DNA adenine methylase, (Dam)[1] (also site-specific DNA-methyltransferase (adenine-specific), EC 2.1.1.72, modification methylase, restriction-modification system) is an enzyme that adds a methyl group to the adenine of the sequence 5'-GATC-3' in newly synthesized DNA.[2][3] Immediately after DNA synthesis, the daughter strand remains unmethylated for a short time.[4] It is an orphan methyltransferase that is not part of a restriction-modification system and regulates gene expression.[5][6][7][8] This enzyme catalyses the following chemical reaction
- S-adenosyl-L-methionine + DNA adenine [math]\displaystyle{ \rightleftharpoons }[/math] S-adenosyl-L-homocysteine + DNA 6-methylaminopurine
This is a large group of enzymes unique to prokaryotes and bacteriophages.[9]
The E. coli DNA adenine methyltransferase enzyme (Dam), is widely used for the chromatin profiling technique DamID, in which the Dam is fused to a DNA-binding protein of interest and expressed as a transgene in a genetically tractable model organism to identify protein binding sites.[10]
Role in mismatch repair of DNA
When DNA polymerase makes an error resulting in a mismatched base-pair or a small insertion or deletion during DNA synthesis, the cell will repair the DNA by a pathway called mismatch repair. However, the cell must be able to differentiate between the template strand and the newly synthesized strand. In some bacteria, DNA strands are methylated by Dam methylase, and therefore, immediately after replication, the DNA will be hemimethylated.[4] A repair enzyme, MutS, binds to mismatches in DNA and recruits MutL, which subsequently activates the endonuclease MutH. MutH binds hemimethylated GATC sites and when activated will selectively cleave the unmethylated daughter strand, allowing helicase and exonucleases to excise the nascent strand in the region surrounding the mismatch.[4][11] The strand is then re-synthesized by DNA polymerase III.
Role in regulation of replication
The firing of the origin of replication (oriC) in bacteria cells is highly controlled to ensure DNA replication occurs only once during each cell division. Part of this can be explained by the slow hydrolysis of ATP by DnaA, a protein that binds to repeats in the oriC to initiate replication. Dam methylase also plays a role because the oriC has 11 5'-GATC-3' sequences (in E. coli). Immediately after DNA replication, the oriC is hemimethylated and sequestered for a period of time. Only after this, the oriC is released and must be fully methylated by Dam methylase before DnaA binding occurs.
Role in regulation of protein expression
Dam also plays a role in the promotion and repression of RNA transcription. In E. coli downstream GATC sequences are methylated, promoting transcription. For example, pyelonephritis-associated pili (PAP) phase variation in uropathogenic E. coli is controlled by Dam through the methylation of the two GATC sites proximal and distal to the PAP promoter.[12] Given its role of protein regulation in E. coli, the Dam methylase gene is nonessential as a knockout of the gene still leaves the bacteria viable.[13] The retainment of viability despite a dam gene knockout is also seen in Salmonella and Aggregatibacter actinomycetemcomitans.[14][15] However, in organisms like Vibrio cholerae and Yersinia pseudotuberculosis, the dam gene is essential for viability.[16] A knockout of the dam gene in Aggregatibacter actinomycetemcomitans resulted in dysregulated levels of the protein, leukotoxin, and also reduced the microbe's ability to invade oral epithelial cells.[15] Additionally a study on Dam methylase deficient Streptococcus mutans, a dental pathogen, revealed the dysregulation of a 103 genes some of which include cariogenic potential.[16]
Structural features
The similarity in the catalytic domains of C5-cytosine methyltransferases and N6 and N4-adenine methyltransferases provided great interest in understanding the basis for functional similarities and dissimilarities. The methyltransferases or methylases are classified into three groups (Groups α, β, and γ) based on the sequential order of certain 9 motifs and the Target Recognition Domain (TRD).[17] Motif I consists of a Gly-X-Gly tripeptide and is referred to as the G-loop and is implicated in the binding of the S-Adenosyl methionine cofactor.[18] Motif II is highly conserved among N4 and N6-adenine methylases and contains a negatively charged amino acid followed by a hydrophobic side chain in the last positions of the β2 strand to bind the AdoMet.[17] Motif III is also implicated in the binding of Adomet. Motif IV is especially important and well known in methylase characterizations. It consists of a diprolyl component and is highly conserved among N6-adenine methyltransferases as the DPPY motif, however, this motif can vary for N4-adenine and C5-cytosine methyltransferases. The DPPY motif has been found to be essential for AdoMet binding.[19] Motifs IV-VIII play a role in the catalytic activity, while motifs 1-III and X play a role in binding of the cofactor. For N6-adenine methylases the sequential order for these motifs is as such: N-terminal - X - I - II - III - TRD - IV - V - VI - VII - VIII - C-terminal and E. coli Dam methylase follows this structural sequence.[17] A 2015 crystallography experiment showed that E. coli Dam methylase was able to bind non-GATC DNA with the same sequence of motifs discussed; the authors posit that the obtained structure could serve as grounds for repression of transcription that is not methylation based.[20]
Orphan bacterial and bacteriophage methylases
Dam methylase is an orphan methyltransferase that is not part of a restriction-modification system but operates independently to regulate gene expression, mismatch repair, and bacterial replication amongst many other functions. This is not the only example of an orphan methyltransferase as there exists the Cell cycle regulated Methyltransferase (CcrM) which methylates 5'-GANTC-'3 hemi-methylated DNA to control the life cycle of Caulobacter crescentus and other related species.[21]
Distinct from their bacterial counterparts, phage orphan methyltransferases also do exist and most notably in the T2, T4, and other T-even bacteriophages that infect E. coli.[5] In a study it was identified that despite sharing any sequence homology, the E. coli and T4 Dam methylase amino acids sequences share sequence identity of up to 64% in four regions of 11 to 33 residues long which suggests a common evolutionary origin for the bacterial and phage methylase genes.[22] The T2 and T4 methylases differ from E. coli Dam methylase in not only their ability to methylate 5-hydroxymethylcytosine but to also methylate non-canonical DNA sites. Despite extensive in vitro characterization of these select few phage orphan methyltransferases their biological purpose is still not clear.[5]
See also
References
- ↑ Brown, Terence (2002). "Chapter 14: Mutation, Repair and Recombination. Section 2.3" (in en). Genomes. Garland Science. ISBN 0-471-25046-5. https://www.ncbi.nlm.nih.gov/books/NBK21114/#A8425.
- ↑ "Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12". Journal of Bacteriology 114 (3): 1143–50. June 1973. doi:10.1128/JB.114.3.1143-1150.1973. PMID 4576399.
- ↑ "Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease". The Journal of Biological Chemistry 254 (4): 1408–13. February 1979. PMID 368070.
- ↑ 4.0 4.1 4.2 "The great GATC: DNA methylation in E. coli". Trends in Genetics 5 (5): 139–143. 1989. doi:10.1016/0168-9525(89)90054-1. PMID 2667217.
- ↑ 5.0 5.1 5.2 "Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence". Applied and Environmental Microbiology 79 (24): 7547–55. December 2013. doi:10.1128/aem.02229-13. PMID 24123737.
- ↑ "Specificity of restriction endonucleases and DNA modification methyltransferases a review (Edition 3)". Gene 92 (1–2): 1–248. August 1990. doi:10.1016/0378-1119(90)90486-B. PMID 2172084.
- ↑ "Restriction enzymes and their isoschizomers". Nucleic Acids Research 18 Suppl: 2331–65. April 1990. doi:10.1093/nar/18.suppl.2331. PMID 2159140.
- ↑ "Structure and mechanism of multifunctional restriction endonucleases". Annual Review of Biochemistry 50: 285–319. 1981. doi:10.1146/annurev.bi.50.070181.001441. PMID 6267988.
- ↑ "The Restriction Enzyme Database". http://rebase.neb.com/rebase/rebase.html.
- ↑ "Dam it's good! DamID profiling of protein-DNA interactions". Wiley Interdisciplinary Reviews. Developmental Biology 5 (1): 25–37. January 2016. doi:10.1002/wdev.205. PMID 26383089.
- ↑ "Dam methylation: coordinating cellular processes". Current Opinion in Microbiology 8 (2): 154–60. April 2005. doi:10.1016/j.mib.2005.02.009. PMID 15802246.
- ↑ "Epigenetic gene regulation in the bacterial world". Microbiology and Molecular Biology Reviews 70 (3): 830–56. September 2006. doi:10.1128/MMBR.00016-06. PMID 16959970.
- ↑ "Characterization of DNA adenine methylation mutants of Escherichia coli K12". Mutation Research 59 (2): 157–65. February 1979. doi:10.1016/0027-5107(79)90153-2. PMID 375073.
- ↑ "DNA methylation-dependent regulation of pef expression in Salmonella typhimurium". Molecular Microbiology 35 (4): 728–42. February 2000. doi:10.1046/j.1365-2958.2000.01743.x. PMID 10692151.
- ↑ 15.0 15.1 "Inactivation of DNA adenine methyltransferase alters virulence factors in Actinobacillus actinomycetemcomitans". Oral Microbiology and Immunology 21 (4): 238–44. August 2006. doi:10.1111/j.1399-302x.2006.00284.x. PMID 16842508.
- ↑ 16.0 16.1 "DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae". Infection and Immunity 69 (12): 7610–5. December 2001. doi:10.1128/iai.69.12.7610-7615.2001. PMID 11705940.
- ↑ 17.0 17.1 17.2 "Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes". Journal of Molecular Biology 253 (4): 618–32. November 1995. doi:10.1006/jmbi.1995.0577. PMID 7473738.
- ↑ "Universal catalytic domain structure of AdoMet-dependent methyltransferases". Journal of Molecular Biology 247 (1): 16–20. March 1995. doi:10.1006/jmbi.1994.0117. PMID 7897657.
- ↑ "Conserved sequence motif DPPY in region IV of the phage T4 Dam DNA-[N-adenine-methyltransferase is important for S-adenosyl-L-methionine binding"]. Nucleic Acids Research 21 (15): 3563–6. July 1993. doi:10.1093/nar/21.15.3563. PMID 16617501.
- ↑ "Structures of Escherichia coli DNA adenine methyltransferase (Dam) in complex with a non-GATC sequence: potential implications for methylation-independent transcriptional repression". Nucleic Acids Research 43 (8): 4296–308. April 2015. doi:10.1093/nar/gkv251. PMID 25845600.
- ↑ "A Caulobacter DNA methyltransferase that functions only in the predivisional cell". Journal of Molecular Biology 235 (2): 472–85. January 1994. doi:10.1006/jmbi.1994.1007. PMID 8289276.
- ↑ "Common evolutionary origin of the phage T4 dam and host Escherichia coli dam DNA-adenine methyltransferase genes". Journal of Bacteriology 164 (2): 932–7. November 1985. doi:10.1128/JB.164.2.932-937.1985. PMID 3902803.
External links
 |