Biology:Halobacterium noricense
| Halobacterium noricense | |
|---|---|
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | Halobacteria
|
| Order: | |
| Family: | |
| Genus: | |
| Species: | H. noricense Fendrihan et al. 2006
|
Halobacterium noricense is a halophilic, rod-shaped microorganism that thrives in environments with salt levels near saturation.[1] Despite the implication of the name, Halobacterium is actually a genus of archaea, not bacteria.[1] H. noricense can be isolated from environments with high salinity such as the Dead Sea and the Great Salt Lake in Utah.[1] Members of the Halobacterium genus are excellent model organisms for DNA replication and transcription due to the stability of their proteins and polymerases when exposed to high temperatures.[2] To be classified in the genus Halobacterium, a microorganism must exhibit a membrane composition consisting of ether-linked phosphoglycerides and glycolipids.[2]
Scientific classification
This organism is a member of the genus Halobacterium and its taxonomic classification is as follows: Archaea, Euryarchaeota, Euryarchaeota, Halobacteria, Halobacteriales, Halobacteriaceae, Halobacterium, Halobacterium noricense.[1] There are currently 19 known halophilic archaeal genera and 57 known species within the genus Halobacterium.[2]
Relatives
Three reported strains Halobacterium salinarium NRC-1, Halobacterium sp. DL1, and Halobacterium salinarium R1 were compared to Halobacterium noricense strain CBA1132.[3] The phylogenetic trees based on Multi-Locus Sequence Typing (MLST) and Average Nucleotide Identity (ANI) indicated that strain CBA1132 and strain DL1 are closely related while strains NRC-1 and R1 are closely related.[3] Multi-Locus Sequence Typing is a technique that uses genomic information to establish evolutionary relationships between bacterial taxa.[4] Average Nucleotide Identity is a genetic method used to compare the similarity between nucleotides of two strains based on the coding regions of their genomes, which has allowed scientists to veer away from traditional methods of classifying prokaryotes based on phenotypic similarities.[5] The defining characteristic between strains CBA1132 and DL1 is that they both contain high GC content in their chromosomes, providing stability in a harsh environment.[6] Other close relatives of H. noricense within the genus Halobacterium include Halobacterium denitrificans, Halobacterium halobium, and Halobacterium volcanii.[2]
Morphology
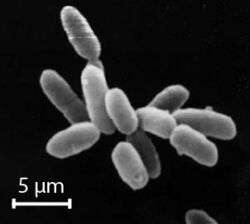
Halobacterium noricense is known to be free living, and it typically appears as red or pink colonies due to the presence of carotenoids and bacterioruberin in their membranes.[3][7] The carotenoids have the ability to absorb light between the wavelengths of 330-600 nm, as determined by light spectroscopy.[2] Typical colony morphology is round with a diameter of 0.4 mm.[2] Under the microscope, they can typically be measured at around 5 μm and appear gram-negative and rod-shaped.[2] H. noricense does not contain the gas vesicles that are present in their close relative, Halobacterium salinarium, which often appear as floating cultures.[2] Halobacterium noricense may occasionally appear as coccus-shaped when grown in liquid broth rather than on solid media.[2]
Discovery
Etymology
Halobacterium noricense is named after Noricum, Austria, which is the location of the salt deposit in which the organism was isolated.[2] The archaeon was discovered in 2004 by a group of scientists led by Claudia Gruber.[2] The group isolated two strains of H. noricense, along with other Halobacterium species including H. salinarium.[2]
Sources
The first two strains (A1 and A2) of Halobacterium noricense were isolated from samples taken out of a salt deposit in Austria.[2] The salt deposit was approximately 400 meters below the surface and is believed to have been formed during the Permian period.[2] To obtain the samples, the researchers used a pre-existing mine to travel below the Earth's surface.[2] They used a core drill to remove cylindrical sections of the salt deposit, which were then taken for sequencing.[2] The deposit retained high salt levels over approximately 250 million years due to the surrounding clay and limestone.[2] These conditions do not allow the salt to escape, which formed an ideal environment for halophilic archaea.[2]
Media
Halobacterium noricense was isolated on ATCC 2185 medium with 250.0 grams of NaCl, 20.0 grams of MgSO4 7H2O, 2.0 grams of KCl, 3.0 grams of yeast extract, 5.0 grams of tryptone, and other compounds required for the isolate's growth.[8] After an incubation period of approximately 2 weeks, red circular colonies appeared.[2] This is the characteristic colony morphology of H. noricense.[2]
Growth Conditions
Halobacterium noricense is known to be a mesophile, where optimum growth temperature is approximately 37 °C with an incubation period of 18 days.[2] It thrives in acidic conditions at pH 5.2-7.0.[2] NaCl concentration between 15-17% has resulted in the highest growth rates in previous studies.[2] It has been found that Halobacterium can survive in high metal concentrations because they are extremely halophilic.[3] This can be achieved through metal resistance, which indicates that the H. noricense strain CBA1132 might also be able to survive in these high metal ion concentrations.[3]
Genome
H. noricense strains A1 and A2 from Gruber et al.[2] had 97.1% similarity to genus Halobacterium through their 16S rDNA sequences.[2] H. noricense genome, strain CBA1132, composed of four contigs containing 3,012,807 base pairs, approximately 3,084 gene coding sequences, and 2,536 genes.[3][9] It has a GC content of approximately 65.95%, and 687 of the genes in the H. noricense genome have unknown functions.[3][9] Metabolism and amino acid transport-related genes make up the largest group of known genes.[9] This group contains 213 known genes.[9] The genus Halobacterium is currently known as monophyletic because their 16S rRNA have less than 80% similarity with their closest relatives, the methanogens.[1]
Sequencing
According to Joint Genome Institute, another complete genome analysis of Halobacterium (strain DL1) species was sequenced using 454 GS FLX, Illumina GAIIx.[10] Halobacterium noricense (strain CBA1132) was recently isolated from solar salt and a complete genomic analysis was performed by researchers from Korea in 2016.[3][9] The researchers extracted the DNA using a QuickGene DNA tissue kit, which uses a membrane with extremely fine pores to collect DNA and nucleic acids.[11] They purified the DNA using the MG Genomic DNA purification kit.[9] Once extracted and purified, the strategy for sequencing the genome was Whole Genome Sequencing by the method of a PacBio RS II system.[9] Lastly, the genome was analyzed and performed by the Rapid Annotation using Subsystem Technology (RAST) server.[3]
Metabolism
According to Gruber et al., Halobacterium noricense cannot ferment glucose, galactose, sucrose, xylose or maltose.[2] It is resistant to many antibiotics, including Vancomycin and Tetracycline, but can be killed by Anisomycin.[2] This organism does not produce the enzymes gelatinase or amylase, so it cannot break down starch or gelatin.[2] H. noricense is a chemoorganotroph and uses aerobic respiration in most environments, except when exposed to L-arginine or nitrate. In these cases, it can function as a facultative anaerobe.[2] It is catalase positive, meaning it has the ability to break down hydrogen peroxide into water and oxygen.[2] The most abundant carbon source found in hypersaline environments is glycerol due to the contribution of the green algae, Dunaliella, to reduce its surrounding osmotic pressure.[12] H. noricense is able to metabolize glycerol through phosphorylation to glycerol 3- phosphate and eventually, into the formation of dihydroxyacetone 5- phosphate (DHAP).[12] NMR spectroscopy, used to locate local magnetic fields around atomic nuclei, revealed during aerobic respiration that 90% of pyruvate that is converted to acetyl Co-A by pyruvate synthase enters the Citric acid cycle while the other 10% is converted to oxaloacetate by biotin carboxylases to later be used in fatty acid degradation.[13]
Ecology
Metagenomic analysis was performed on concentrated biomass from the last Dead Sea bloom and compared with hundreds of liters of brine (pH 6), revealing that the bloom was less diverse from brine.[14] The Dead Sea is located on the borders of Israel and the Jordan River where its depth is around 300 m.[14] The Dead Sea contains 1.98M Mg2+, 1.54M Na+, and 0.08M (1%) Br− making the waters unique and the ecosystem harsh.[14]
Samples were collected from the Dead Sea in 1992 at Ein Gedi 310 station during bloom season.[14] The cells were centrifuged and a reddish cell pellet was embedded in agarose plugs.[14] DNA was extracted from the plugs and cloned into pCC1fos vector to construct two fosmid libraries, which contain DNA from bacterial F-plasmids.[14]
BAC-end sequences were performed on each library for further analysis, and the sequences were scanned for vector contamination and removed by BLASTing.[14] The read length was 734 bp for the 1992 library.[14]
PCR 16S rRNA gene amplification was carried out and was used to construct a tree to calculate bootstrap values from a total of 714 sequence positions.[14] Although halophiles are diverse, analysis revealed that most rRNAs had around 93% similarity to sequences in GenBank.[14] H. noricense had a 95% similarity in the 1992 bloom.[14] When the samples were compared to the fosmid library, some were over 88% similar to other known halophilic bacterial species.[14] This indicates that these halophiles are specifically adapted to the extreme salinity of the Dead Sea.[14]
There are also studies in the field of astrobiology regarding the possibility of Halobacterium on Mars.[15] Similarly to the Dead Sea, any water located on the Martian surface would be a brine with an extremely high salt concentration.[15] Therefore, microbial life on Mars would require adaptations similar to those of Halobacterium.[15]
Significance
Halobacterium noricense has many applications that can benefit humans and industries including drug delivery, UV protection, and the unique characteristic of bacteriorhodopsin to be able to be isolated outside of its environment.[16] H. noricense produces a high concentration of menaquinones (fat soluble vitamin K2) that can be used as a micelle to deliver drugs to specific places in the body.[16] According to Nimptsch K, the presence of menaquinones can also reduce the risk of malignant cancer.[16] Fermented foods are also found to have high levels of menaquinones due to the presence of bacteria, especially in cheeses.[17] H. noricense requires high salt concentrations and is currently being explored to enhance the process of fermentation.[18] H. noricense is also catalase positive, meaning it can break down reactive oxygen species (ROS), like hydrogen peroxide into harmless substances such as water.[18] Not only does it produce enzymes to protect itself against ROS, but it contains a pigment, bacterioruberin, that allows H. noricense to tolerate gamma and UV radiation.[18] Further research into bacterioruberin can lead to bioactive compounds with anticancer characteristics.[18] Lastly, bacteriorhodopsin (also protects cells from UV light), a light proton pump, has allowed scientists to apply it to electronics and optics. Its mechanism involves capturing light and creating a proton gradient to produce chemical energy. Some practical uses include motion detection, holographic storage, and nanotechnology.[19]
References
- ↑ 1.0 1.1 1.2 1.3 1.4 "Extremely halophilic archaea and the issue of long-term microbial survival". Re/Views in Environmental Science and Bio/Technology 5 (2–3): 203–218. August 2006. doi:10.1007/s11157-006-0007-y. PMID 21984879.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 2.18 2.19 2.20 2.21 2.22 2.23 2.24 2.25 2.26 2.27 2.28 2.29 "Halobacterium noricense sp. nov., an archaeal isolate from a bore core of an alpine Permian salt deposit, classification of Halobacterium sp. NRC-1 as a strain of H. salinarum and emended description of H. salinarum". Extremophiles 8 (6): 431–9. December 2004. doi:10.1007/s00792-004-0403-6. PMID 15290323.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Ki Lim, Seul; Kim, Joon Yong; Seon Song, Hye; Kwon, Min-Sung; Lee, Join; Jun Oh, Young; Nam, Young-Do; Seo, Myung-Ji et al. (2016-05-09). "Genomic Analysis of the Extremely Halophilic Archaeon Halobacterium noricense CBA1132 Isolated from Solar Salt That Is an Essential Material for Fermented Foods". Journal of Microbiology and Biotechnology 26 (8): 1375–1382. doi:10.4014/jmb.1603.03010. PMID 27160574. https://www.researchgate.net/publication/302910769.
- ↑ Maiden, M. C.; Jansen Van Rensburg, M. J.; Bray, J. E.; Earle, S. G.; Ford, S. A.; Jolley, K. A.; McCarthy, N. D. (2013). "MLST revisited: the gene-by-gene approach to bacterial genomics". Nature Reviews. Microbiology 11 (10): 728–736. doi:10.1038/nrmicro3093. PMID 23979428.
- ↑ Zhang W., Pengcheng D., Han Z. et al. (2014) Whole-genome sequence comparison as a method for improving bacterial species definition. J. Gen. Appl. Microbiol., 60, 75–78.
- ↑ Pfeiffer, F., Schuster, S.C., Broicher, A., Falb, M., Palm, P., Rodewald, K., et al., Evolution in the laboratory: The genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1, Genomics, 2008, 91(4):335-346.
- ↑ Fendrihan, S.; Legat, A.; Pfaffenhuemer, M.; Gruber, C.; Weidler, G.; Gerbl, F.; Stan-Lotter, H. (2006). "Extremely halophilic archaea and the issue of long-term microbial survival". Re/Views in Environmental Science and Bio/Technology (Online) 5 (2–3): 203–218. doi:10.1007/s11157-006-0007-y. PMID 21984879.
- ↑ "ATCC medium: 2185 Halobacterium NRC-1 medium". American Type Culture Collection (ATCC). https://www.atcc.org/~/media/2F6B035C311C45C8A56814C2DED11A75.ashx.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 "Genomic Analysis of the Extremely Halophilic Archaeon Halobacterium noricense CBA1132 Isolated from Solar Salt That Is an Essential Material for Fermented Foods". Journal of Microbiology and Biotechnology 26 (8): 1375–82. August 2016. doi:10.4014/jmb.1603.03010. PMID 27160574.
- ↑ "IMG". https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2506783047.
- ↑ "Wako-Chem". http://www.wako-chem.co.jp/english/labchem/product/me/QG/pdf/DT-S_v4.pdf.
- ↑ 12.0 12.1 Borowitzka, Lesley Joyce; Kessly, David Stuart; Brown, Austin Duncan (1977-05-01). "The salt relations of Dunaliella" (in en). Archives of Microbiology 113 (1–2): 131–138. doi:10.1007/BF00428592. ISSN 0302-8933. PMID 19000.
- ↑ Ghosh, M.; Sonawat, Haripalsingh M. (1998-11-01). "Kreb's TCA cycle in Halobacterium salinarum investigated by 13C nuclear magnetic resonance spectroscopy" (in en). Extremophiles 2 (4): 427–433. doi:10.1007/s007920050088. ISSN 1431-0651. PMID 9827332.
- ↑ 14.00 14.01 14.02 14.03 14.04 14.05 14.06 14.07 14.08 14.09 14.10 14.11 14.12 Bodaker, Idan; Sharon, Itai; Suzuki, Marcelino T; Feingersch, Roi; Shmoish, Michael; Andreishcheva, Ekaterina; Sogin, Mitchell L; Rosenberg, Mira et al. (2009-12-24). "Comparative community genomics in the Dead Sea: an increasingly extreme environment" (in En). The ISME Journal 4 (3): 399–407. doi:10.1038/ismej.2009.141. ISSN 1751-7362. PMID 20033072.
- ↑ 15.0 15.1 15.2 Landis, Geoffrey A. (2001). "Martian Water". Astrobiology 1 (2): 161–164. doi:10.1089/153110701753198927. PMID 12467119. https://zenodo.org/record/896700.
- ↑ 16.0 16.1 16.2 Nimptsch, Katharina; Rohrmann, Sabine; Kaaks, Rudolf; Linseisen, Jakob (2010-03-24). "Dietary vitamin K intake in relation to cancer incidence and mortality: results from the Heidelberg cohort of the European Prospective Investigation into Cancer and Nutrition (EPIC-Heidelberg)" (in en). The American Journal of Clinical Nutrition 91 (5): 1348–1358. doi:10.3945/ajcn.2009.28691. ISSN 0002-9165. PMID 20335553.
- ↑ Hojo, K.; Watanabe, R.; Mori, T.; Taketomo, N. (September 2007). "Quantitative measurement of tetrahydromenaquinone-9 in cheese fermented by propionibacteria". Journal of Dairy Science 90 (9): 4078–4083. doi:10.3168/jds.2006-892. ISSN 1525-3198. PMID 17699024.
- ↑ 18.0 18.1 18.2 18.3 Gontia-Mishra, Iti; Sapre, Swapnil; Tiwari, Sharad (2017). "Diversity of halophilic bacteria and actinobacteria from India and their biotechnological applications". Indian Journal of Geo-Marine Sciences 46 (8): 1575–1587. ISSN 0975-1033. http://nopr.niscair.res.in/handle/123456789/42523.
- ↑ Oren, Aharon (July 2017). "Industrial and environmental applications of halophilic microorganisms". Environmental Technology 31 (8–9): 825–834. doi:10.1080/09593330903370026. PMID 20662374.
Further reading
- "Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1". Genomics 91 (4): 335–46. April 2008. doi:10.1016/j.ygeno.2008.01.001. PMID 18313895.
- "Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation". Extremophiles 9 (3): 219–27. June 2005. doi:10.1007/s00792-005-0437-4. PMID 15844015. https://hal-mnhn.archives-ouvertes.fr/mnhn-02862359/file/2005_Kottemann_Kish_NRC1%20physiological%20response%20to%20desiccation%20and%20gamma%20radiation.pdf.
- "Gas vesicles isolated from Halobacterium cells by lysis in hypotonic solution are structurally weakened". FEMS Microbiology Letters 252 (2): 337–41. November 2005. doi:10.1016/j.femsle.2005.09.017. PMID 16213677.
External links
Wikidata ☰ Q16117914 entry
 |