Biology:Halobacterium
| Halobacterium | |
|---|---|
| |
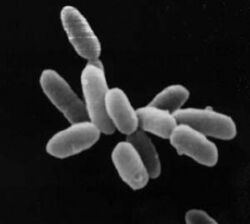
| Halobacterium sp. strain NRC-1, each cell about 5 μm in length | |
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | Halobacteria
|
| Order: | |
| Family: | |
| Genus: | Halobacterium (Elazari-Volcani 1940) Elazari-Volcani 1957 non Schoop 1935
|
| Type species | |
| Halobacterium salinarum (Harrison & Kennedy 1922) Elazari-Volcani 1957
| |
| Species | |
| |
| Synonyms | |
| |
Halobacterium (common abbreviation Hbt.) is a genus in the family Halobacteriaceae.[1]
The genus Halobacterium ("salt" or "ocean bacterium") consists of several species of Archaea with an aerobic metabolism which requires an environment with a high concentration of salt; many of their proteins will not function in low-salt environments. They grow on amino acids in their aerobic conditions. Their cell walls are also quite different from those of bacteria, as ordinary lipoprotein membranes fail in high salt concentrations. In shape, they may be either rods or cocci, and in color, either red or purple. They reproduce using binary fission (by constriction), and are motile. Halobacterium grows best in a 42 °C environment. The genome of an unspecified Halobacterium species, sequenced by Shiladitya DasSarma, comprises 2,571,010 bp (base pairs) of DNA compiled into three circular strands: one large chromosome with 2,014,239 bp, and two smaller ones with 191,346 and 365,425 bp. This species, called Halobacterium sp. NRC-1, has been extensively used for postgenomic analysis. Halobacterium species can be found in the Great Salt Lake, the Dead Sea, Lake Magadi, and any other waters with high salt concentration. Purple Halobacterium species owe their color to bacteriorhodopsin, a light-sensitive protein which provides chemical energy for the cell by using sunlight to pump protons out of the cell. The resulting proton gradient across the cell membrane is used to drive the synthesis of the energy carrier ATP. Thus, when these protons flow back in, they are used in the synthesis of ATP (this proton flow can be emulated with a decrease in pH outside the cell, causing a flow of H+ ions). The bacteriorhodopsin protein is chemically very similar to the light-detecting pigment rhodopsin, found in the vertebrate retina.
Species of Halobacterium
Phylogeny
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN)[2] and National Center for Biotechnology Information (NCBI).[3]
| 16S rRNA based LTP_08_2023[4][5][6] | 53 marker proteins based GTDB 08-RS214[7][8][9] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
Synonyms
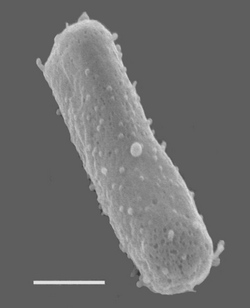
Size bar = 270 nm
- Halobacterium cutirubrum > Halobacterium salinarum
- Halobacterium denitrificans > Haloferax denitrificans
- Halobacterium distributum > Halorubrum distributum
- Halobacterium halobium > Halobacterium salinarum
- Halobacterium lacusprofundi > Halorubrum lacusprofundi
- Halobacterium mediterranei > Haloferax mediterranei
- Halobacterium pharaonis > Natronomonas pharaonis
- Halobacterium piscisalsi > Halobacterium salinarum
- Halobacterium saccharovorum > Halorubrum saccharovorum
- Halobacterium sodomense > Halorubrum sodomense
- Halobacterium trapanicum > Halorubrum trapanicum
- Halobacterium vallismortis > Haloarcula vallismortis
- Halobacterium volcanii > Haloferax volcanii
Genome structure
The Halobacterium NRC-1 genome is 2,571,010 bp compiled into three circular replicons. More specifically, it is divided into one large chromosome with 2,014,239 bp and two small replicons pNRC100 (191,346 bp) and pNRC200 (365,425 bp). While much smaller than the large chromosome, the two plasmids account for most of the 91 insertion sequences and include genes for a DNA polymerase, seven transcription factors, genes in potassium and phosphate uptake, and cell division. The genome was discovered to contain a high G+C content at 67.9% on the large chromosome and 57.9% and 59.2% on the two plasmids. The genome also contained 91 insertion sequence elements constituting 12 families, including 29 on pNRC100, 40 on pNRC200, and 22 on the large chromosome. This helps explain the genetic plasticity that has been observed in Halobacterium. Of the archaea, halobacteria are viewed as being involved in the most lateral genetics (gene transfer between domains) and a proof that this transfer does take place.
Cell structure and metabolism
Halobacterium species are rod-shaped and enveloped by a single lipid bilayer membrane surrounded by an S-layer made from the cell-surface glycoprotein. They grow on amino acids in aerobic conditions. Although Halobacterium NRC-1 contains genes for glucose degradation, as well as genes for enzymes of a fatty acid oxidation pathway, it does not seem able to use these as energy sources. Though the cytoplasm retains an osmotic equilibrium with the hypersaline environment, the cell maintains a high potassium concentration using many active transporters.
Many Halobacterium species possess proteinaceous organelles called gas vesicles.
Ecology
Halobacteria can be found in highly saline lakes such as the Great Salt Lake, the Dead Sea, and Lake Magadi. Halobacterium can be identified in bodies of water by the light-detecting pigment bacteriorhodopsin, which not only provides the archaeon with chemical energy, but adds to its reddish hue as well. An optimal temperature for growth has been observed at 37 °C.
Halobacterium may be a candidate for a life form present on Mars. One of the problems associated with the survival on Mars is the destructive ultraviolet light. These microorganisms develop a thin crust of salt that can moderate some of the ultraviolet light. Sodium chloride is the most common salt and chloride salts are opaque to short-wave ultraviolet. Their photosynthetic pigment, bacteriorhodopsin, is actually opaque to the longer-wavelength ultraviolet (its red color). In addition, Halobacterium makes pigments called bacterioruberins that are thought to protect cells from damage by ultraviolet light. The obstacle they need to overcome is being able to grow at a low temperature during a presumably short time when a pool of water could be liquid.
Applications
Food Industry
There is potential for Halobacterium species to be used in the food industry.[10] Some examples of uses can include the production of Beta-Carotene, a pigment in halophilic bacteria that contributes to their red coloration, is used in the food industry as a natural food dye. Halophiles also produce degradative enzymes such as lipases, amylases, proteases, and xylanases that are used in various food processing methods. Notable applications of these enzymes include enhancing the fermentation process of salty foods, improving dough quality for the baking of breads, and contributing to the production of coffee.[10][11]
Bioremediation
Many species of halophilic bacteria produce exopolysaccharides (EPS) which are used industrially as bioremediation agents. Biosurfactants are also released by many halophilic bacteria and these amphiphilic compounds have been used for soil remediation. Many halophiles are highly tolerant of heavy metals making them potentially useful in the bioremediation of xenobiotic compounds and heavy metals that are released into the environment from mining and metal plating. Halophiles contribute to the bioremediation of contaminants by converting xenobiotics into less toxic compounds.[11] Some Halobacterium species have been shown to be effective in the bioremediation of pollutants including aliphatic hydrocarbons, such as those found in crude oil; and aromatic hydrocarbons such as 4-hydroxybenzoic acid, a contaminant in some high salinity industrial runoffs.[citation needed]
Pharmaceuticals
Some strains of Halobacterium, including Halobacterium salinarum, are being explored for medical applications of their radiation-resistance mechanisms. Bacterioruberin is a carotenoid pigment found in Halobacterium which decreases the bacteria’s sensitivity to γ-radiation and UV radiation.[12]
It has been shown in knockout studies, that the absence of bacterioruberin increases the sensitivity of the bacterium to oxidative DNA-damaging agents. Hydrogen peroxide, for example, reacts with bacteroruberin which prevents the production of reactive oxygen species, and thus protects the bacterium by reducing the oxidative capacity of the DNA-damaging agent.[13]
H. salinarum also exhibits high intracellular concentrations of potassium chloride which has also been shown to confer radiation resistance. Halobacterium are also being explored for the pharmaceutical applications of bioactive compounds they produce, including anticancer agents, antimicrobial biosurfactancts, and antimicrobial metabolites.[12]
Significance and applications
Halobacteria are halophilic microorganisms that are currently being studied for their uses in scientific research and biotechnology. For instance, genomic sequencing of the Halobacterium species NRC-1 revealed their use of eukaryotic-like RNA polymerase II and translational machinery that are related to Escherichia coli and other Gram-negative bacteria. In addition, they possess genes for DNA replication, repair, and recombination that are similar to those present in bacteriophages, yeasts, and bacteria. The ability of this Halobacterium species to be easily cultured and genetically modified allows it to be used as a model organism in biological studies.[14] Furthermore, Halobacterium NRC-1 have also been employed as a potential vector for delivering vaccines. In particular, they produce gas vesicles that can be genetically engineered to display specific epitopes. Additionally, the gas vesicles demonstrate the ability to function as natural adjuvants to help evoke stronger immune responses. Because of the requirement of Halobacteria for a high-salt environment, the preparation of these gas vesicles is inexpensive and efficient, needing only tap water for their isolation.[15]
Halobacteria also contain a protein called Bacteriorhodopsins which are light-driven proton pumps found on the cell membrane. Although most proteins in halophiles need high salt concentrations for proper structure and functioning, this protein has shown potential to be used for biotechnological purposes because of its stability even outside of these extreme environments. Bacteriorhodopsins isolated from Halobacterium salinarum have been especially studied for their applications in electronics and optics. Particularly, bacteriorhodopsins have been used in holographic storage, optical switching, motion detection, and nanotechnology. Although numerous uses of this protein have been presented, there are yet to be any high-scale commercial applications established at this time.[16]
Recombination and mating
UV irradiation of Halobacterium sp. strain NRC-1 induces several gene products employed in homologous recombination.[17] For instance, a homolog of the rad51/recA gene, which plays a key role in recombination, is induced 7-fold by UV. Homologous recombination may rescue stalled replication forks, and/or facilitate recombinational repair of DNA damage.[17] In its natural habitat, homologous recombination is likely induced by the UV irradiation in sunlight.
Halobacterium volcanii has a distinctive mating system in which cytoplasmic bridges between cells appear to be used for transfer of DNA from one cell to another.[18] In wild populations of Halorubrum, genetic exchange and recombination occur frequently.[19] This exchange may be a primitive form of sexual interaction, similar to the more well studied bacterial transformation that is also a process for transferring DNA between cells leading to homologous recombinational repair of DNA damage.[citation needed]
See also
Further reading
Scientific journals
- DasSarma, S., B.R. Berquist, J.A. Coker, P. DasSarma, J.A. Müller. 2006. Post-genomics of the model haloarchaeon Halobacterium sp. NRC-1. Saline Systems 2:3.
- Judicial, Commission of the International Committee on Systematics of Prokaryotes (2005). "The nomenclatural types of the orders Acholeplasmatales, Halanaerobiales, Halobacteriales, Methanobacteriales, Methanococcales, Methanomicrobiales, Planctomycetales, Prochlorales, Sulfolobales, Thermococcales, Thermoproteales and Verrucomicrobiales are the genera Acholeplasma, Halanaerobium, Halobacterium, Methanobacterium, Methanococcus, Methanomicrobium, Planctomyces, Prochloron, Sulfolobus, Thermococcus, Thermoproteus and Verrucomicrobium, respectively. Opinion 79". Int. J. Syst. Evol. Microbiol. 55 (Pt 1): 517–518. doi:10.1099/ijs.0.63548-0. PMID 15653928.
- "International Committee on Systematic Bacteriology Subcommittee on the taxonomy of Halobacteriaceae. Minutes of the meetings, 16 August 1999, Sydney, Australia". Int. J. Syst. Evol. Microbiol. 50 (3): 1405–1407. 2000. doi:10.1099/00207713-50-3-1405. PMID 10843089.
Scientific books
- DasSarma, S. 2004. Genome sequence of an extremely halophilic archaeon, in Microbial Genomes, pp. 383–399, C.M. Fraser, T. Read, and K.E. Nelson (eds.), Humana Press, Inc., Totowa, NJ.
- Lynn Margulis, Karlene V.Schwartz, Five Kingdoms. An Illustrated Guide to the Phyla of Life on Earth (W.H.Freeman, San Francisco, 1982) pp. 36–37
- Gibbons, NE (1974). "Family V. Halobacteriaceae fam. nov.". in RE Buchanan. Bergey's Manual of Determinative Bacteriology (8th ed.). Baltimore: The Williams & Wilkins Co..
- Elazari-Volcani, B (1957). "Genus XII. Halobacterium Elazari-Volcani, 1940". in RS Breed. Bergey's Manual of Determinative Bacteriology (7th ed.). Baltimore: The Williams & Wilkins Co.. pp. 207–212. https://archive.org/details/bergeysmanualofd1957amer.
- Elazari-Volcani, B (1940). Studies on the microflora of the Dead Sea. Doctoral dissertation, Hebrew University, Jerusalem. pp. 1–116 and i–xiii.
References
- ↑ See the NCBI webpage on Halobacterium. Data extracted from the "NCBI taxonomy resources". National Center for Biotechnology Information. http://ftp.ncbi.nih.gov/pub/taxonomy/.
- ↑ J.P. Euzéby. "Halobacterium". List of Prokaryotic names with Standing in Nomenclature (LPSN). https://lpsn.dsmz.de/genus/halobacterium.
- ↑ Sayers. "Halobacterium". National Center for Biotechnology Information (NCBI) taxonomy database. https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=2239&lvl=3&keep=1&srchmode=1&unlock.
- ↑ "The LTP". https://imedea.uib-csic.es/mmg/ltp/#LTP.
- ↑ "LTP_all tree in newick format". https://imedea.uib-csic.es/mmg/ltp/wp-content/uploads/ltp/LTP_all_08_2023.ntree.
- ↑ "LTP_08_2023 Release Notes". https://imedea.uib-csic.es/mmg/ltp/wp-content/uploads/ltp/LTP_08_2023_release_notes.pdf.
- ↑ "GTDB release 08-RS214". https://gtdb.ecogenomic.org/about#4%7C.
- ↑ "ar53_r214.sp_label". https://data.gtdb.ecogenomic.org/releases/release214/214.0/auxillary_files/ar53_r214.sp_labels.tree.
- ↑ "Taxon History". https://gtdb.ecogenomic.org/taxon_history/.
- ↑ 10.0 10.1 Paterson, Russell; Lima, Nelson (2017). "Bioprospecting". Topics in Biodiversity and Convervation. Topics in Biodiversity and Conservation 16: 84–91. doi:10.1007/978-3-319-47935-4. ISBN 978-3-319-47933-0.
- ↑ 11.0 11.1 Gontia-Mishra, Iti; Sapre, Swapnil; Tiwari, Sharad (August 2017). "Diversity of halophilic bacteria and actinobacteria from India and their biotechnological applications". Indian Journal of Geo-Marine Sciences 46 (8): 1575–1587. http://nopr.niscair.res.in/handle/123456789/42523. Retrieved 8 October 2017.
- ↑ 12.0 12.1 Jung, Kwang-Woo; Lim, Sangyong; Bahn, Yong-Sun (30 June 2017). "Microbial radiation-resistance mechanisms". Journal of Microbiology 55 (7): 499–507. doi:10.1007/s12275-017-7242-5. PMID 28664512.
- ↑ SHAHMOHAMMADI, HAMID REZA; ASGARANI, EZAT; TERATO, HIROAKI; SAITO, TAKESHI; OHYAMA, YOSHIHIKO; GEKKO, KUNIHIKO; YAMAMOTO, OSAMU; IDE, HIROSHI (1998). "Protective Roles of Bacterioruberin and Intracellular KCl in the Resistance of Halobacterium salinarium against DNA-damaging Agents.". Journal of Radiation Research 39 (4): 251–262. doi:10.1269/jrr.39.251. PMID 10196780. Bibcode: 1998JRadR..39..251S.
- ↑ Ng, W. V.; Kennedy, S. P.; Mahairas, G. G.; Berquist, B.; Pan, M.; Shukla, H. D.; Lasky, S. R.; Baliga, N. S. et al. (3 October 2000). "Genome sequence of Halobacterium species NRC-1". Proceedings of the National Academy of Sciences 97 (22): 12176–12181. doi:10.1073/pnas.190337797. PMID 11016950. Bibcode: 2000PNAS...9712176N.
- ↑ Stuart, Elizabeth S.; Morshed, Fazeela; Sremac, Marinko; DasSarma, Shiladitya (15 June 2001). "Antigen presentation using novel particulate organelles from halophilic archaea". Journal of Biotechnology 88 (2): 119–128. doi:10.1016/S0168-1656(01)00267-X. PMID 11403846.
- ↑ Oren, Aharon (July 2010). "Industrial and environmental applications of halophilic microorganisms". Environmental Technology 31 (8–9): 825–834. doi:10.1080/09593330903370026. PMID 20662374.
- ↑ 17.0 17.1 "UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1". Saline Syst. 1: 3. 2005. doi:10.1186/1746-1448-1-3. PMID 16176594.
- ↑ "The mechanism of DNA transfer in the mating system of an archaebacterium". Science 245 (4924): 1387–9. 1989. doi:10.1126/science.2818746. PMID 2818746. Bibcode: 1989Sci...245.1387R.
- ↑ "Frequent recombination in a saltern population of Halorubrum". Science 306 (5703): 1928–9. 2004. doi:10.1126/science.1103289. PMID 15591201. Bibcode: 2004Sci...306.1928P.
External links
Wikidata ☰ Q139014 entry
 |
