Biology:Siderocalin
 Generic protein structure example |
Siderocalin (Scn), lipocalin-2, NGAL, 24p3 is a mammalian lipocalin-type protein that can prevent iron acquisition by pathogenic bacteria by binding siderophores, which are iron-binding chelators made by microorganisms.[1][2] Iron serves as a key nutrient in host-pathogen interactions, and pathogens can acquire iron from the host organism via synthesis and release siderophores such as enterobactin.[3] Siderocalin is a part of the mammalian defence mechanism and acts as an antibacterial agent.[1][4][5][6][7] Crystallographic studies of Scn demonstrated that it includes a calyx, a ligand-binding domain that is lined with polar cationic groups.[8] Central to the siderophore/siderocalin recognition mechanism are hybrid electrostatic/cation-pi interactions.[5][9] To evade the host defences, pathogens evolved to produce structurally varied siderophores that would not be recognized by siderocalin, allowing the bacteria to acquire iron.[1]
Iron requirements of host organisms
Organisms require iron for a variety of chemical reactions.[10] Although iron can be found throughout the biosphere, free ferric iron forms insoluble hydroxides at physiological pH, limiting its accessibility in aerobic conditions to living organisms.[10][11] In order to preserve homeostasis, organisms have evolved specific protein networks, with proteins and receptors translated in accordance with intracellular iron levels.[10][12] Export and import are supplemented by a cycling process between the ferrous Fe(II) available in the reducing environment of the cell, and ferric Fe(III) found primarily under aerobic conditions.[13] [14] The iron acquisition mechanisms of pathogenic bacteria demonstrate the role of iron as a key component at the interface between pathogens and hosts.[13][14]
Lipocalin family of iron binding proteins
The lipocalin family of binding proteins are produced by the immune system and sequester ferric siderophore complexes from the siderophore receptors of bacteria.[15] [16] The lipocalin family of binding proteins typically have a conserved eight-stranded β-barrel fold with a calyx binding site,[16][17] which are lined with positively charged amino acid residues, allowing for binding interactions with siderophores.[citation needed]
Clinical significance
Mycobacterial infections
The lipocalin siderocalin is found in neutrophil granules, uterine secretions, and at particularly high levels in serum during bacterial infection.[4] Upon infection, pathogens use siderophores to capture iron from the host organism.[18] This strategy is, however, complicated by the human protein siderocalin, which can sequester siderophores, and prevent their use by pathogenic bacteria as iron delivery agents.[19] This effect has been demonstrated by studies with siderocalin-knock-out mice, which are more sensitive to infections under iron-limiting conditions.[4][5]
Mycobacterial virulence
Siderophores are iron chelators, allowing organisms to acquire iron from their environment. In the case of pathogens, iron can be acquired from the host organism.[20] Siderophores and ferric iron can associate to form stable complexes.[10][21][22] Siderophores bind iron using a variety of ligands, most commonly as α-hydroxycarboxylates (e.g. citrate), catecholates, and hydroxamates.[5][10][23] [24] As a defence mechanism, siderocalin can substitute ferric bis-catechol complexes (formed under physiological conditions) with a third catechol, in order to achieve a hexacoordinate ferric complex, resulting in higher affinity binding.[5][18][25]
As a mediator of mammalian iron transport
Mammalian siderophores, specifically catechols, can be found in the human gut and in siderophores, such as enterobactin, and serve as iron-binding moieties.[5][26] Catechol resembling molecules can act as iron ligands in the cell and in systematic circulation, allowing siderocalin to bind to the iron-catechol complex.[27] Catechols can be bound by siderocalin, in the form of free ligands, or in the iron complex.[28] 24p3 is a vertebrate lipocalin-2 receptor which allows for import of the ferric siderophore complex into mammalian cells.[27] During kidney embryogenesis, siderocalin mediated iron transport occurs, as iron concentration has to be highly controlled in order to restrict inflammation.[4][11] Following secretion by neutrophils, siderocalin can bind to pathogenic siderophores, such as bacillibactin, and prevent siderophore trafficking.[29] Siderocalin has been linked with various cellular processes apart from iron transport, including apoptosis, cellular differentiation, tumorigenesis, and metastasis.[10][30]
Structure
The avian orthologs of siderocalin (Q83 and Ex-FABP) and NGAL (neutrophil gelatinase-associated lipocalin-2) contain calyces with positively charged lysine and arginine side chains.[8][30][31][32][33] These side chains interact via cation-pi and coulombic interactions with the negatively charged siderophores that contain aromatic catecholate groups.[10][30] Crystallographic studies of siderocalin have shown that the ligand binding domain of Scn, known as the calyx, is shallow and broad, and is lined with polar cationic groups from the three positively charged residues of Arg81, Lys125, and Lys134.[5][8][34] Scn can also bind non-ferric complexes and has been identified as a potential transporter for heavy actinide ions. Scn crystal structures containing heavy metals (thorium, plutonium, americium, curium, and californium) have been obtained.[35][36] Scn has been found as a monomer, homo-dimer, or trimer in human plasma.[5] The siderocalin fold is exceptionally stable.[4][5] The calyx is structurally stable and rigid, and conformational change does not typically occur upon a change in pH, ionic strength, or ligand binding.[5]
Binding pocket
The structural stability of the calyx has been attributed to the three binding pockets within the calyx that sterically limit which ligands are compatible with siderocalin.[5][8] The Scn calyx can accommodate three aromatic rings of the catecholate moieties, in the three available binding pockets.[5][28] Solid-state and solution structural results demonstrated that bacteria-derived enterobactin is bound to the binding pocket of Scn, allowing for Scn to be involved in the acute immune response to bacterial infection.[5][21] One method by which pathogens can circumvent immunity mechanisms is by modifying the siderophore chemical structure to prevent interaction with Scn.[24] One example is the addition of glucose molecules to the enterobactin backbone of salmochelin (C-glucosylated enterobactin) in order to increase the hydrophilicity and bulkiness of a siderophore and inhibit binding to Scn.[24][37]
Binding interactions
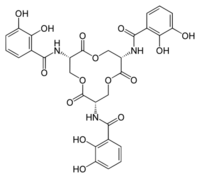
Siderophores are typically bound to siderocalin with subnanomolar affinities, and interact with siderocalin specifically.[10][25] The Kd value of the siderocalin/siderophore interaction, measured by fluorescence quenching (Kd= 0.4 nM), indicates that siderocalin can capture siderophores with high affinity.[31][38] This Kd value is similar to that of the FepA bacterial receptor (Kd= 0.3 nM).[5] Siderophore/siderocalin binding is directed by electrostatic interactions.[5][38] Specifically, the mechanism involves hybrid electrostatic and cation-pi interactions in the positively charged protein calyx.[25] The siderophore is positioned in the centre of the siderocalin calyx, and is associated with multiple direct polar interactions.[25] Structural analysis of the siderocalin/siderophore interaction has shown that the siderophore is accompanied by a poor and diffuse quality of electron density, with the majority of the ligand exposed to the solvent when the siderophore is fit in the calyx.[5][6] Siderocalin typically does not bind hydroxamate-based siderophores because these substrates do not have the necessary aromatic electronic structure for cation-pi interactions.[5][25] In order to acquire iron in the presence of siderocalin, pathogenic bacteria utilize several siderophores that do not bind to siderocalin, or structurally modify siderophores to inhibit siderocalin binding.[5][39] Siderocalin can bind soluble siderophores of mycobacteria, including carboxymycobactins.[5][6] In vivo studies have shown that the binding interactions between carboxymycobactin and siderocalin serve to protect the host organism from mycobacterial infections, with siderocalin inhibiting mycobacterial iron acquisition.[5][28][40] Siderocalin can sequester ferric carboxymycobactins by employing a polyspecific recognition mechanism.[5] The siderophore/siderocalin recognition mechanism primarily involves hybrid electrostatic/cation-pi interactions.[5][9][11] The fatty acid tails of carboxymycobactin reside in a ‘tail-in’ or ‘tail-out’ conformation within pocket 2.[5] The ‘tail-in’ conformation of the fatty acid chain lengths introduces a significant interaction between the calyx and the ligand, increasing the affinity of the siderocalin calyx and carboxymycobactin.[5] The fatty acid tails of short lengths have a correspondingly less favorable binding to siderocalin, and cannot maintain the necessary interaction with the binding pocket.[5] Since lipocalin-2 cannot bind the long fatty acid chain carboxymycobactins of mycobacteria, it is apparent that a number of pathogens have evolved to avoid the activity of lipocalin-2.[41]
Recognition mechanism
Electrostatic interaction play a key role in the recognition mechanism of siderophores by siderocalin.[1] The binding of the siderophore and the siderocalin binding pocket is primarily directed by cation-pi interactions, with the positively charged binding pocket of siderocalin attracting the negatively charged complex.[1] A structural factor involved in the siderocalin mediated recognition mechanism of phenolate/catecholate-type siderophores includes a backbone linker which allows for siderocalin to interact with different phenolate/catecholate siderophores.[4][42] While siderocalin recognition is minimally affected by the substitution of different metals, methylating the three catecholate rings of enterobactin can impede the recognition of siderocalin.[5][34][38][43] A strategy used by pathogens to overcome immune response is the production of siderophores that will not be recognized by siderocalin.[19][44] For example, siderocalin cannot recognize the siderophores of the C-glucosylated analog of enterobactin, as the donor groups are glycosylated, introducing steric interactions at the position 5-carbons of the catechol groups.[1][24]
History
The requirement for iron by humans and pathogens has been known for many years.[10] The link between iron and mycobactins, iron-chelating growth factors from mycobacteria, was first made in the 1960s.[5] At the time, interest was growing in resolving an application of mycobactins as target molecules for a rational anti-tuberculosis agent.[5][45] Experiments in the 1960s and 1970s demonstrated that iron deficiency in mycobacteria was the cause of 'anaemic’ cells.[46] The majority of the genes and systems necessary for high affinity iron acquisition have been identified in pathogenic and saprophytic mycobacteria.[5] These genes encode proteins for iron storage, uptake of ferric-siderophores, and heme.[5][47] Humans have evolved a defense for siderophore-mediated iron acquisition by developing siderocalin. To combat this, various pathogens have evolved siderophores that can evade siderocalin recognition.[5] Siderocalin has been shown to bind to siderophores and inhibit iron acquisition, and prevent the growth of Mycobacterium tuberculosis in extracellular cultures; however, the effect of siderocalin on this pathogen within macrophages remains unclear.[24][31]
See also
- LCN2
- LCN1
- Animal pathogens
- Mycobacteria
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 van Eldik, R.; Hubbard, C. D. (2009). Advances in Inorganic Chemistry Volume 61 (1st ed.). London, U.K.: Elsevier. pp. 237–239. ISBN 9780123750334. https://books.google.com/books?id=Q2bSgQijqmUC&q=siderocalin&pg=PA238. Retrieved 16 February 2015.
- ↑ "Siderocalin/Lcn2/NGAL/24p3 does not drive apoptosis through gentisic acid mediated iron withdrawal in hematopoietic cell lines". PLOS ONE 7 (8): e43696. 2012. doi:10.1371/journal.pone.0043696. PMID 22928018. Bibcode: 2012PLoSO...743696C.
- ↑ "The role of iron in the immune response to bacterial infection". Immunologic Research 50 (1): 1–9. May 2011. doi:10.1007/s12026-010-8199-1. PMID 21161695.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 "NGAL-Siderocalin in kidney disease". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1823 (9): 1451–8. Sep 2012. doi:10.1016/j.bbamcr.2012.06.014. PMID 22728330.
- ↑ 5.00 5.01 5.02 5.03 5.04 5.05 5.06 5.07 5.08 5.09 5.10 5.11 5.12 5.13 5.14 5.15 5.16 5.17 5.18 5.19 5.20 5.21 5.22 5.23 5.24 5.25 5.26 5.27 5.28 5.29 5.30 Byers, B. R. (2013). Iron Acquisition by the Genus Mycobacterium. SpringerBriefs in Molecular Science. Springer. pp. 1–88. doi:10.1007/978-3-319-00303-0. ISBN 978-3-319-00303-0.
- ↑ 6.0 6.1 6.2 "Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration". Structure 13 (1): 29–41. Jan 2005. doi:10.1016/j.str.2004.10.009. PMID 15642259.
- ↑ Sige l, A.; Sigel, H.; Sige l, R. K. O. (2013). Interrelations between Essential Metal Ions and Human Diseases. Heidelberg, Germany: Springer. pp. 282–283. ISBN 9789400774995. https://books.google.com/books?id=6OIlBAAAQBAJ&q=siderocalin&pg=PA283. Retrieved 14 February 2015.
- ↑ 8.0 8.1 8.2 8.3 "Siderocalins: Siderophore binding proteins evolved for primary pathogen host defense". Current Opinion in Chemical Biology 17 (2): 150–7. Apr 2013. doi:10.1016/j.cbpa.2012.11.014. PMID 23265976.
- ↑ 9.0 9.1 "Anthrax pathogen evades the mammalian immune system through stealth siderophore production". Proceedings of the National Academy of Sciences of the United States of America 103 (49): 18499–503. Dec 2006. doi:10.1073/pnas.0607055103. PMID 17132740. Bibcode: 2006PNAS..10318499A.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 10.7 10.8 "Mammalian siderophores, siderophore-binding lipocalins, and the labile iron pool". The Journal of Biological Chemistry 287 (17): 13524–31. Apr 2012. doi:10.1074/jbc.R111.311829. PMID 22389496.
- ↑ 11.0 11.1 11.2 Chakraborty, R.; Braun, V.; Hantke, K.; Cornelis, P. (2013). Iron Uptake in Bacteria with Emphasis on E. coli and Pseudomonas. SpringerBriefs in Biometals. pp. 31–66. ISBN 978-94-007-6087-5.
- ↑ "Systemic iron homeostasis". Physiological Reviews 93 (4): 1721–41. Oct 2013. doi:10.1152/physrev.00008.2013. PMID 24137020.
- ↑ 13.0 13.1 "Host-pathogen interactions: the role of iron". The Journal of Nutrition 137 (5): 1341–4. May 2007. doi:10.1093/jn/137.5.1341. PMID 17449603.
- ↑ 14.0 14.1 "The battle for iron between bacterial pathogens and their vertebrate hosts". PLOS Pathogens 6 (8): e1000949. 2010. doi:10.1371/journal.ppat.1000949. PMID 20711357.
- ↑ "Microbial iron acquisition: marine and terrestrial siderophores". Chemical Reviews 109 (10): 4580–95. Oct 2009. doi:10.1021/cr9002787. PMID 19772347.
- ↑ 16.0 16.1 "The lipocalin protein family: structure and function". The Biochemical Journal 318 (1): 1–14. Aug 1996. doi:10.1042/bj3180001. PMID 8761444.
- ↑ "Structure of the thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug". Proceedings of the National Academy of Sciences of the United States of America 94 (22): 11845–50. Oct 1997. doi:10.1073/pnas.94.22.11845. PMID 9342325. Bibcode: 1997PNAS...9411845F.
- ↑ 18.0 18.1 "Siderophore-based iron acquisition and pathogen control". Microbiology and Molecular Biology Reviews 71 (3): 413–51. Sep 2007. doi:10.1128/MMBR.00012-07. PMID 17804665.
- ↑ 19.0 19.1 "Siderocalin outwits the coordination chemistry of vibriobactin, a siderophore of Vibrio cholerae". ACS Chemical Biology 8 (9): 1882–7. Sep 2013. doi:10.1021/cb4002552. PMID 23755875.
- ↑ "Molecular strategies of microbial iron assimilation: from high-affinity complexes to cofactor assembly systems". Metallomics 5 (1): 15–28. Jan 2013. doi:10.1039/C2MT20193C. PMID 23192658.
- ↑ 21.0 21.1 "The siderocalin/enterobactin interaction: a link between mammalian immunity and bacterial iron transport". Journal of the American Chemical Society 130 (34): 11524–34. Aug 2008. doi:10.1021/ja803524w. PMID 18680288.
- ↑ "Gram-positive siderophore-shuttle with iron-exchange from Fe-siderophore to apo-siderophore by Bacillus cereus YxeB". Proceedings of the National Academy of Sciences of the United States of America 110 (34): 13821–6. Aug 2013. doi:10.1073/pnas.1304235110. PMID 23924612. Bibcode: 2013PNAS..11013821F.
- ↑ "Iron chelation equilibria, redox, and siderophore activity of a saccharide platform ferrichrome analogue". Inorganic Chemistry 46 (20): 8362–71. Oct 2007. doi:10.1021/ic070158l. PMID 17824601. http://people.duke.edu/~jmh36/papers/suraj%20sucrose%20paper%202.pdf.
- ↑ 24.0 24.1 24.2 24.3 24.4 Yehuda, S.; Mostofsky, D. I. (2010). Iron Deficiency and Overload: From Basic Biology to Clinical Medicine. New York, N.Y.: Humana Press. pp. 66–69. ISBN 9781934115220. https://books.google.com/books?id=gyByd18ZdRcC&q=siderocalin&pg=PA68. Retrieved 14 February 2015.
- ↑ 25.0 25.1 25.2 25.3 25.4 "The role of electrostatics in siderophore recognition by the immunoprotein Siderocalin". Journal of the American Chemical Society 130 (51): 17584–92. Dec 2008. doi:10.1021/ja8074665. PMID 19053425.
- ↑ "Iron-Binding Catechols and Virulence in Escherichia coli". Infection and Immunity 7 (3): 445–56. Mar 1973. doi:10.1128/IAI.7.3.445-456.1973. PMID 16558077.
- ↑ 27.0 27.1 Anderson, G. J.; McLaren, G. D. (2012). Iron Physiology and Pathophysiology in Humans. New York, N.Y.: Springer. pp. 237–239, 658. ISBN 9781603274845. https://books.google.com/books?id=wtmeZX2Ku6AC&q=lipocalin2&pg=PA235. Retrieved 14 February 2015.
- ↑ 28.0 28.1 28.2 "Iron traffics in circulation bound to a siderocalin (Ngal)-catechol complex". Nature Chemical Biology 6 (8): 602–9. Aug 2010. doi:10.1038/nchembio.402. PMID 20581821.
- ↑ Bergman, N. H. (2011). Bacillus anthracis and Anthrax. Hoboken, N.J.: Wiley. pp. Chapter 7. ISBN 9781118148082. https://books.google.com/books?id=ZvoB2V8mEmgC&q=siderocalin&pg=PT136. Retrieved 14 February 2015.
- ↑ 30.0 30.1 30.2 "Galline Ex-FABP is an antibacterial siderocalin and a lysophosphatidic acid sensor functioning through dual ligand specificities". Structure 19 (12): 1796–806. Dec 2011. doi:10.1016/j.str.2011.09.019. PMID 22153502.
- ↑ 31.0 31.1 31.2 Ashton Acton, Q. (2012). Advances in Serine Research and Application (2012: ScholarlyBrief ed.). Atlanta, Georgia: ScholarlyEditions. pp. 42–43. ISBN 9781481614276. https://books.google.com/books?id=7ZchzymxiVMC&q=siderocalin&pg=PR3. Retrieved 14 February 2015.
- ↑ Thongboonkerd, V. (2007). Proteomics of Human Body Fluids: Principles, Methods, and Applications. Totowa, N.J: Humana Press. pp. 338–339. ISBN 9781597454322. https://books.google.com/books?id=6SkzhODYFxQC&q=lipocalin+2&pg=PA338. Retrieved 16 February 2015.
- ↑ "Siderocalins: siderophore-binding proteins of the innate immune system". Biometals 22 (4): 557–64. Aug 2009. doi:10.1007/s10534-009-9207-6. PMID 19184458. https://labs.fhcrc.org/strong/Clifton.Biometals.09.pdf.
- ↑ 34.0 34.1 "Immune interference in Mycobacterium tuberculosis intracellular iron acquisition through siderocalin recognition of carboxymycobactins". ACS Chemical Biology 6 (12): 1327–31. Dec 2011. doi:10.1021/cb200331g. PMID 21978368.
- ↑ Deblonde, Gauthier J.-P.; Sturzbecher-Hoehne, Manuel; Rupert, Peter B.; Dahlia, An D.; Illy Marie-Claire; Ralston, Corie Y.; Brabec, Jiri; de Jong, Wide A. et al. (September 2017). "Chelation and stabilization of berkelium in oxidation state +IV". Nature Chemistry 9 (9): 843–849. doi:10.1038/nchem.2759. ISSN 1755-4349. PMID 28837177. Bibcode: 2017NatCh...9..843D. https://escholarship.org/content/qt9zn3q96n/qt9zn3q96n.pdf?t=ow5vil.
- ↑ Captain, Ilya; Deblonde, Gauthier J.-P.; Rupert, Peter B.; An, Dahlia D.; Illy, Marie-Claire; Rostan, Emeline; Ralston, Corie Y.; Strong, Roland K. et al. (2016-11-21). "Engineered Recognition of Tetravalent Zirconium and Thorium by Chelator–Protein Systems: Toward Flexible Radiotherapy and Imaging Platforms". Inorganic Chemistry 55 (22): 11930–11936. doi:10.1021/acs.inorgchem.6b02041. ISSN 0020-1669. PMID 27802058. https://escholarship.org/uc/item/2nx8r6pz.
- ↑ Alvarez, M. V. (2007). Isolation, Structure and Detection of Salmochelins: Novel Siderophores in Enterobacteria. Göttingen, Germany: Cuvillier Verlag. pp. 29–34. ISBN 9783867271097. https://books.google.com/books?id=86y1twJ75vIC&q=siderophores. Retrieved 14 February 2015.
- ↑ 38.0 38.1 38.2 "Microbial evasion of the immune system: structural modifications of enterobactin impair siderocalin recognition". Journal of the American Chemical Society 128 (34): 10998–9. Aug 2006. doi:10.1021/ja062476+. PMID 16925397.
- ↑ "Ebola virus antibody prevalence in dogs and human risk". Emerging Infectious Diseases 11 (3): 385–90. Mar 2005. doi:10.3201/eid1103.040981. PMID 15757552.
- ↑ Åkerström, B. (2006). Lipocalins. Austin, Texas: Landes Bioscience. p. 92. ISBN 9781587062971.
- ↑ Kidd, S. P. (2011). Stress Response in Pathogenic Bacteria, Volume 19 of Advances in Molecular and Cellular Microbiology. Wallingford, U.K.: CABI. pp. 287–290. ISBN 9781845937775. https://books.google.com/books?id=ixDDwBUuL_IC&q=lipocalin+2. Retrieved 14 February 2015.
- ↑ Strong, R. K.; Akerstrom, B.; Borregaard, N.; Flower, D. R.; Salier, J.-P. (Eds.). "Siderocalins". Fred Hutchinson Cancer Research Center. https://labs.fhcrc.org/strong/Akerstrom08Strong.pdf.
- ↑ "Enterobactin protonation and iron release: structural characterization of the salicylate coordination shift in ferric enterobactin". Journal of the American Chemical Society 128 (27): 8920–31. Jul 2006. doi:10.1021/ja062046j. PMID 16819888.
- ↑ "Microbial iron transport via a siderophore shuttle: a membrane ion transport paradigm". Proceedings of the National Academy of Sciences of the United States of America 97 (20): 10691–6. Sep 2000. doi:10.1073/pnas.200318797. PMID 10995480. Bibcode: 2000PNAS...9710691S.
- ↑ "Iron-binding compounds of Mycobacterium avium, M. intracellulare, M. scrofulaceum, and mycobactin-dependent M. paratuberculosis and M. avium". Journal of Bacteriology 153 (3): 1138–46. Mar 1983. doi:10.1128/JB.153.3.1138-1146.1983. PMID 6826517.
- ↑ Jamison, D. T.; Breman, J. G.; Measham, A. R.; Alleyne, G.; Claeson, M.; Evans, D. B.; Jha, P.; Mills, A. et al. (2006). Disease Control Priorities in Developing Countries (2nd ed.). Washington, D.C.: World Bank. pp. Chapter 16. ISBN 978-0-8213-6179-5. https://www.ncbi.nlm.nih.gov/books/NBK11728/. Retrieved 16 February 2015.
- ↑ "Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans". Frontiers in Cellular and Infection Microbiology 3 (80): 80. 2013. doi:10.3389/fcimb.2013.00080. PMID 24312900.
 |