Biology:MACPF
| MAC/Perforin domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | MACPF | ||||||||
| Pfam | PF01823 | ||||||||
| Pfam clan | CDC | ||||||||
| InterPro | IPR001862 | ||||||||
| SMART | MACPF | ||||||||
| PROSITE | PDOC00251 | ||||||||
| TCDB | 1.C.39 | ||||||||
| OPM superfamily | 168 | ||||||||
| OPM protein | 6h04 | ||||||||
| Membranome | 233 | ||||||||
| |||||||||
The Membrane Attack Complex/Perforin (MACPF) superfamily, sometimes referred to as the MACPF/CDC superfamily,[1] is named after a domain that is common to the membrane attack complex (MAC) proteins of the complement system (C6, C7, C8α, C8β and C9) and perforin (PF). Members of this protein family are pore-forming toxins (PFTs).[2] In eukaryotes, MACPF proteins play a role in immunity and development.[3]
Archetypal members of the family are complement C9 and perforin, both of which function in human immunity.[4] C9 functions by punching holes in the membranes of Gram-negative bacteria. Perforin is released by cytotoxic T cells and lyses virally infected and transformed cells. In addition, perforin permits delivery of cytotoxic proteases called granzymes that cause cell death.[5] Deficiency of either protein can result in human disease.[6][7] Structural studies reveal that MACPF domains are related to cholesterol-dependent cytolysins (CDCs), a family of pore forming toxins previously thought to only exist in bacteria.[8][9]
Families
As of early 2016, there are three families belonging to the MACPF superfamily:
- 1.C.12 - The Thiol-activated Cholesterol-dependent Cytolysin (CDC) Family
- 1.C.39 - The Membrane Attack Complex/Perforin (MACPF) Family
- 1.C.97 - The Pleurotolysin Pore-forming (Pleurotolysin) Family
Membrane Attack Complex/Perforin (MACPF) Family
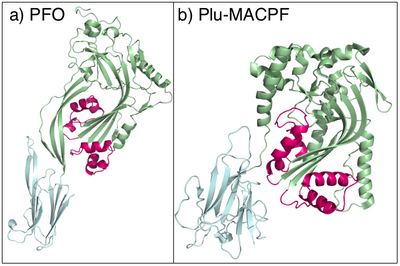
Proteins containing MACPF domains play key roles in vertebrate immunity, embryonic development, and neural-cell migration.[10] The ninth component of complement and perforin form oligomeric pores that lyse bacteria and kill virus-infected cells, respectively. The crystal structure of a bacterial MACPF protein, Plu-MACPF from Photorhabdus luminescens was determined (PDB: 2QP2).[11] The MACPF domain is structurally similar to pore-forming cholesterol-dependent cytolysins from gram-positive bacteria, suggesting that MACPF proteins create pores and disrupt cell membranes similar to cytolysin. A representative list of proteins belonging to the MACPF family can be found in the Transporter Classification Database.
Biological roles of MACPF domain containing proteins
Many proteins belonging to the MACPF superfamily play key roles in plant and animal immunity.
Complement proteins C6-C9 all contain a MACPF domain and assemble into the membrane attack complex. C6, C7 and C8β appear to be non-lytic and function as scaffold proteins within the MAC. In contrast both C8α and C9 are capable of lysing cells. The final stage of MAC formation involves polymerisation of C9 into a large pore that punches a hole in the outer membrane of gram-negative bacteria.
Perforin is stored in granules within cytotoxic T-cells and is responsible for killing virally infected and transformed cells. Perforin functions via two distinct mechanisms. Firstly, like C9, high concentrations of perforin can form pores that lyse cells. Secondly, perforin permits delivery of the cytotoxic granzymes A and B into target cells. Once delivered, granzymes are able to induce apoptosis and cause target cell death.[5][12]
The plant protein CAD1 (TC# 1.C.39.11.3) functions in the plant immune response to bacterial infection.[13][14]
The sea anemone Actineria villosa uses a MACPF (AvTX-60A; TC# 1.C.39.10.1)protein as a lethal toxin.[15]
MACPF proteins are also important for the invasion of the Malarial parasite into the mosquito host and the liver.[16][17]
Not all MACPF proteins function in defence or attack. For example, astrotactin-1 (TC# 9.B.87.3.1) is involved in neural cell migration in mammals and apextrin (TC# 1.C.39.7.4) is involved in sea urchin (Heliocidaris erythrogramma) development.[18][19] Drosophila Torso-like protein (TC# 1.C.39.15.1), which controls embryonic patterning,[20] also contains a MACPF domain.[8] Its function is implicated in a receptor tyrosine kinase signaling pathway that specifies differentiation and terminal cell fate.
Functionally uncharacterised MACPF proteins are sporadically distributed in bacteria. Several species of Chlamydia contain MACPF proteins.[21] The insect pathogenic bacteria Photorhabdus luminescens also contains a MACPF protein, however, this molecule appears non-lytic.[8]
Structure and mechanism
The X-ray crystal structure of Plu-MACPF, a protein from the insect pathogenic enterobacteria Photorhabdus luminescens has been determined (figure 1).[5] These data reveal that the MACPF domain is homologous to pore forming cholesterol dependent cytolysins (CDC's) from gram-positive pathogenic bacteria such as Clostridium perfringens (which causes gas gangrene). The amino acid sequence identity between the two families is extremely low, and the relationship is not detectable using conventional sequence based data mining techniques.[8]
It is suggested that MACPF proteins and CDCs form pores in the same way (figure 1).[8] Specifically it is hypothesised that MACPF proteins oligomerise to form a large circular pore (figure 2). A concerted conformational change within each monomer then results in two α-helical regions unwinding to form four amphipathic β-strands that span the membrane of the target cell.[8] Like CDC's MACPF proteins are thus β-pore forming toxins that act like a molecular hole punch.
Other crystal structures for members of the MACPF superfamily can be found in RCSB: i.e., 3KK7, 3QOS, 3QQH, 3RD7, 3OJY
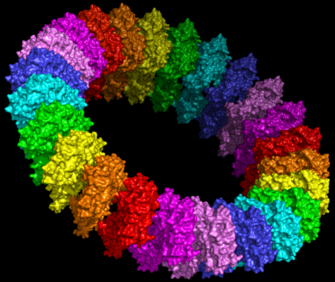 Figure 2: Molecular model of the pre-pore form of a MACPF protein based upon the structure of pneunolysin.[23] |
Control of MACPF proteins
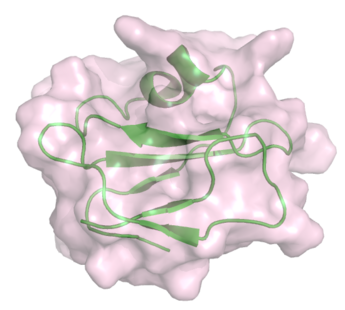
Complement regulatory proteins such as CD59 function as MAC inhibitors and prevent inappropriate activity of complement against self cells (Figure 3). Biochemical studies have revealed the peptide sequences in C8α and C9 that bind to CD59.[24][25] Analysis of the MACPF domain structures reveals that these sequences map to the second cluster of helices that unfurl to span the membrane. It is therefore suggested that CD59 directly inhibits the MAC by interfering with conformational change in one of the membrane spanning regions.[8]
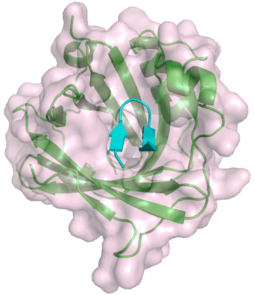
Other proteins that bind to the MAC include C8γ. This protein belongs to the lipocalin family and interacts with C8α. The binding site on C8α is known, however, the precise role of C8γ in the MAC remains to be understood.[26][27]
Role in human disease
Deficiency of C9, or other components of the MAC results in an increased susceptibility to diseases caused by gram-negative bacteria such as meningococcal meningitis.[29] Overactivity of MACPF proteins can also cause disease. Most notably, deficiency of the MAC inhibitor CD59 results in an overactivity of complement and Paroxysmal nocturnal hemoglobinuria.[30]
Perforin deficiency results in the commonly fatal disorder familial hemophagocytic lymphohistiocytosis (FHL or HLH).[6] This disease is characterised by an overactivation of lymphocytes which results in cytokine mediated organ damage.[31]
The MACPF protein DBCCR1 may function as a tumor suppressor in bladder cancer.[3][32]
Human proteins containing this domain
C6; C7; C8A; C8B; C9; FAM5B; FAM5C; MPEG1; PRF1
References
- ↑ Gilbert, Robert J. C.; Mikelj, Miha; Dalla Serra, Mauro; Froelich, Christopher J.; Anderluh, Gregor (2013-06-01). "Effects of MACPF/CDC proteins on lipid membranes". Cellular and Molecular Life Sciences 70 (12): 2083–2098. doi:10.1007/s00018-012-1153-8. ISSN 1420-9071. PMID 22983385.
- ↑ "Assembly of macromolecular pores by immune defense systems". Curr. Opin. Cell Biol. 3 (4): 710–6. 1991. doi:10.1016/0955-0674(91)90045-Z. PMID 1722985.
- ↑ 3.0 3.1 Rosado, Carlos J.; Kondos, Stephanie; Bull, Tara E.; Kuiper, Michael J.; Law, Ruby H. P.; Buckle, Ashley M.; Voskoboinik, Ilia; Bird, Phillip I. et al. (2008-09-01). "The MACPF/CDC family of pore-forming toxins" (in en). Cellular Microbiology 10 (9): 1765–1774. doi:10.1111/j.1462-5822.2008.01191.x. ISSN 1462-5822. PMID 18564372.
- ↑ "Structural/functional similarity between proteins involved in complement- and cytotoxic T-lymphocyte-mediated cytolysis". Nature 322 (6082): 831–4. 1986. doi:10.1038/322831a0. PMID 2427956. Bibcode: 1986Natur.322..831T.
- ↑ 5.0 5.1 "Perforin-mediated target-cell death and immune homeostasis". Nat. Rev. Immunol. 6 (12): 940–52. 2006. doi:10.1038/nri1983. PMID 17124515.
- ↑ 6.0 6.1 "Perforin activity and immune homeostasis: the common A91V polymorphism in perforin results in both presynaptic and postsynaptic defects in function". Blood 110 (4): 1184–90. 2007. doi:10.1182/blood-2007-02-072850. PMID 17475905.
- ↑ "The human complement C9 gene: identification of two mutations causing deficiency and revision of the gene structure". J. Immunol. 158 (10): 5043–9. 1997. doi:10.4049/jimmunol.158.10.5043. PMID 9144525.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 "A Common Fold Mediates Vertebrate Defense and Bacterial Attack". Science 317 (5844): 1548–51. 2007. doi:10.1126/science.1144706. PMID 17717151. Bibcode: 2007Sci...317.1548R.
- ↑ Michael A. Hadders; Dennis X. Beringer; Piet Gros (2007). "Structure of C8-MACPF Reveals Mechanism of Membrane Attack in Complement Immune Defense". Science 317 (5844): 1552–1554. doi:10.1126/science.1147103. PMID 17872444.
- ↑ Anderluh, Gregor; Lakey, Jeremy H. (2008-10-01). "Disparate proteins use similar architectures to damage membranes". Trends in Biochemical Sciences 33 (10): 482–490. doi:10.1016/j.tibs.2008.07.004. ISSN 0968-0004. PMID 18778941.
- ↑ Rosado, Carlos J.; Buckle, Ashley M.; Law, Ruby H. P.; Butcher, Rebecca E.; Kan, Wan-Ting; Bird, Catherina H.; Ung, Kheng; Browne, Kylie A. et al. (2007-09-14). "A common fold mediates vertebrate defense and bacterial attack". Science 317 (5844): 1548–1551. doi:10.1126/science.1144706. ISSN 1095-9203. PMID 17717151. Bibcode: 2007Sci...317.1548R.
- ↑ "The major human and mouse granzymes are structurally and functionally divergent". J. Cell Biol. 175 (4): 619–30. 2006. doi:10.1083/jcb.200606073. PMID 17116752.
- ↑ "The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain". Plant Cell Physiol. 46 (6): 902–12. 2005. doi:10.1093/pcp/pci095. PMID 15799997.
- ↑ Tsutsui, Tomokazu; Morita-Yamamuro, Chizuko; Asada, Yutaka; Minami, Eiichi; Shibuya, Naoto; Ikeda, Akira; Yamaguchi, Junji (2006-09-01). "Salicylic acid and a chitin elicitor both control expression of the CAD1 gene involved in the plant immunity of Arabidopsis". Bioscience, Biotechnology, and Biochemistry 70 (9): 2042–2048. doi:10.1271/bbb.50700. ISSN 0916-8451. PMID 16960394.
- ↑ "A new membrane-attack complex/perforin (MACPF) domain lethal toxin from the nematocyst venom of the Okinawan sea anemone Actineria villosa". Toxicon 43 (2): 225–8. 2004. doi:10.1016/j.toxicon.2003.11.017. PMID 15019483.
- ↑ "Essential role of membrane-attack protein in malarial transmission to mosquito host". Proc. Natl. Acad. Sci. U.S.A. 101 (46): 16310–5. 2004. doi:10.1073/pnas.0406187101. PMID 15520375.
- ↑ "A Plasmodium sporozoite protein with a membrane attack complex domain is required for breaching the liver sinusoidal cell layer prior to hepatocyte infection". Cell. Microbiol. 7 (2): 199–208. 2005. doi:10.1111/j.1462-5822.2004.00447.x. PMID 15659064.
- ↑ "CNS gene encoding astrotactin, which supports neuronal migration along glial fibers". Science 272 (5260): 417–9. 1996. doi:10.1126/science.272.5260.417. PMID 8602532. Bibcode: 1996Sci...272..417Z.
- ↑ "Apextrin, a novel extracellular protein associated with larval ectoderm evolution in Heliocidaris erythrogramma". Dev. Biol. 211 (1): 77–87. 1999. doi:10.1006/dbio.1999.9283. PMID 10373306.
- ↑ "Terminal pattern elements in Drosophila embryo induced by the torso-like protein". Nature 367 (6465): 741–5. 1994. doi:10.1038/367741a0. PMID 8107870. Bibcode: 1994Natur.367..741M.
- ↑ Ponting CP (1999). "Chlamydial homologues of the MACPF (MAC/perforin) domain". Curr. Biol. 9 (24): R911–3. doi:10.1016/S0960-9822(00)80102-5. PMID 10608922.
- ↑ "Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form". Cell 89 (5): 685–92. 1997. doi:10.1016/S0092-8674(00)80251-2. PMID 9182756.
- ↑ "Structural basis of pore formation by the bacterial toxin pneumolysin". Cell 121 (2): 247–56. 2005. doi:10.1016/j.cell.2005.02.033. PMID 15851031.
- ↑ "Defining the CD59-C9 binding interaction". J. Biol. Chem. 281 (37): 27398–404. 2006. doi:10.1074/jbc.M603690200. PMID 16844690.
- ↑ "Identity of the segment of human complement C8 recognized by complement regulatory protein CD59". J. Biol. Chem. 270 (34): 19723–8. 1995. doi:10.1074/jbc.270.8.3483. PMID 7544344.
- ↑ "Expression and characterization of recombinant subunits of human complement component C8: further analysis of the function of C8 alpha and C8 gamma". J. Immunol. 161 (1): 311–8. 1998. doi:10.4049/jimmunol.161.1.311. PMID 9647238.
- ↑ 27.0 27.1 "Crystal structure of complement protein C8gamma in complex with a peptide containing the C8gamma binding site on C8alpha: Implications for C8gamma ligand binding". Molecular Immunology 45 (3): 750–6. 2007. doi:10.1016/j.molimm.2007.06.359. PMID 17692377.
- ↑ "Three-dimensional solution structure of the extracellular region of the complement regulatory protein CD59, a new cell-surface protein domain related to snake venom neurotoxins". Biochemistry 33 (15): 4471–82. 1994. doi:10.1021/bi00181a006. PMID 7512825.
- ↑ "Nonsense mutation in exon 4 of human complement C9 gene is the major cause of Japanese complement C9 deficiency". Hum. Genet. 102 (6): 605–10. 1998. doi:10.1007/s004390050749. PMID 9703418.
- ↑ Walport, Mark J. (2001). "Complement. First of two parts". N. Engl. J. Med. 344 (14): 1058–66. doi:10.1056/NEJM200104053441406. PMID 11287977.
- ↑ "Hemophagocytic lymphohistiocytosis: diagnosis, pathophysiology, treatment, and future perspectives". Ann. Med. 38 (1): 20–31. 2006. doi:10.1080/07853890500465189. PMID 16448985.
- ↑ "DBCCR1 mediates death in cultured bladder tumor cells". Oncogene 23 (1): 82–90. 2004. doi:10.1038/sj.onc.1206642. PMID 14712213.
 |