Biology:Acetyl—CoA synthetase
| Acetate-CoA ligase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 6.2.1.1 | ||||||||
| CAS number | 9012-31-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Acetyl—CoA synthetase (ACS) or Acetate—CoA ligase is an enzyme (EC 6.2.1.1) involved in metabolism of acetate. It is in the ligase class of enzymes, meaning that it catalyzes the formation of a new chemical bond between two large molecules.
Reaction
The two molecules joined together that make up Acetyl CoA synthetase are acetate and coenzyme A (CoA). The complete reaction with all the substrates and products included is:
- ATP + Acetate + CoA <=> AMP + Pyrophosphate + Acetyl-CoA [1]
Once acetyl-CoA is formed it can be used in the TCA cycle in aerobic respiration to produce energy and electron carriers. This is an alternate method to starting the cycle, as the more common way is producing acetyl-CoA from pyruvate through the pyruvate dehydrogenase complex. The enzyme’s activity takes place in the mitochondrial matrix so that the products are in the proper place to be used in the following metabolic steps.[2] Acetyl Co-A can also be used in fatty acid synthesis, and a common function of the synthetase is to produce acetyl Co-A for this purpose.[3]
The reaction catalyzed by acetyl-CoA synthetase takes place in two steps. First, AMP must be bound by the enzyme to cause a conformational change in the active site, which allows the reaction to take place. The active site is referred to as the A-cluster.[4] A crucial lysine residue must be present in the active site to catalyze the first reaction where Co-A is bound. Co-A then rotates in the active site into the position where acetate can covalently bind to CoA. The covalent bond is formed between the sulfur atom in Co-A and the central carbon atom of acetate.[5]
The ACS1 form of acetyl-CoA synthetase is encoded by the gene facA, which is activated by acetate and deactivated by glucose.[6]
Structure
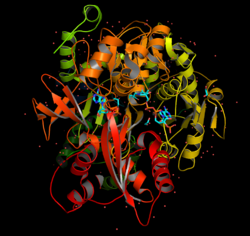
The three dimensional structure of the asymmetric ACS (RCSB PDB ID number: 1PG3) reveals that it is composed of two subunits. Each subunit is then composed primarily of two domains. The larger N-terminal domain is composed of 517 residues, while the smaller C-terminal domain is composed of 130 residues.[7] Each subunit has an active site where the ligands are held. The crystallized structure of ACS was determined with CoA and Adenosine- 5′-propylphosphate bound to the enzyme. The reason for using Adenosine- 5′-propylphosphate is that it is an ATP competitive inhibitor which prevents any conformational changes to the enzyme. The adenine ring of AMP/ATP is held in a hydrophobic pocket create by residues Ile (512) and Trp (413).[7]
The source for the crystallized structure is the organism Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720). The gene for ACS was then transfected into Escherichia coli BL21(DE3) for expression. During chromatography in the process to isolate the enzyme, the subunits came out individually and the total structure was determined separately.[7] The method used to determine the structure was X-ray diffraction with a resolution of 2.3 angstroms. The unit cell values and angles are provided in the following table:

| Length (Å) | Angle (°) |
|---|---|
| a= 59.981 | α= 90.00 |
| b= 143.160 | β= 91.57 |
| c= 71.934 | γ= 90.00 |
Function
The role of the ACS enzyme is to combine acetate and CoA to form acetyl CoA, however its significance is much larger. The most well known function of the product from this enzymatic reaction is the use of Acetyl-CoA in the role of the TCA cycle as well as in the production of fatty acid. This enzyme is vital to the action of histone acetylation as well as gene regulation.[9] The effect this acetylation has is far reaching in mammals. It has been shown that downregulation of the acs gene in the hippocampal region of mice results in lower levels of histone acetylation, but also impairs the long-term spatial memory of the animal. This result points to a link between cellular metabolism, gene regulation and cognitive function.[9] This enzyme has shown to be an interesting biomarker for the presence of tumors in colorectal carcinomas. When the gene is present, the cells are able to take in acetate as a food source to convert it to Acetyl-CoA during stressed conditions. In the cases of advanced carcinoma tumors, the genes for this enzyme were down regulated and indicated a poor 5-year survival rate.[10] Expression of the enzyme has also been linked to the development of metastatic tumor nodes, leading to a poor survival rate in patients with renal cell carcinomas.[11]
Regulation
The activity of the enzyme is controlled in several ways. The essential lysine residue in the active site plays an important role in regulation of activity. The lysine molecule can be deacetylated by another class of enzyme called sirtuins. In mammals, the cytoplasmic-nuclear synthetase (AceCS1) is activated by SIRT1 while the mitochondrial synthetase (AceCS2) is activated by SIRT3. This action increases activity of this enzyme.[2] The exact location of the lysine residue varies between species, occurring at Lys-642 in humans, but is always present in the active site of the enzyme.[12] Since there is an essential allosteric change that occurs with the binding of an AMP molecule, the presence of AMP can contribute to regulation of the enzyme. Concentration of AMP must be high enough so that it can bind in the allosteric binding site and allow the other substrates to enter the active site. Also, copper ions deactivate acetyl Co-A synthetase by occupying the proximal site of the A-cluster active site, which prevents the enzyme from accepting a methyl group to participate in the Wood-Ljungdahl Pathway.[4] The presence of all the reactants in the proper concentration is also needed for proper functioning as in all enzymes. Acetyl—CoA synthetase is also produced when it is needed for fatty acid synthesis, but, under normal conditions, the gene is inactive and has certain transcriptional factors that activate transcription when necessary.[3] In addition to sirtuins, protein deacetylase (AcuC) also can modify acetyl—CoA synthetase at a lysine residue. However, unlike sirtuins, AcuC does not require NAD+ as a cosubstrate.[13]
Biological Significance
The role of the ACS enzyme is to combine acetate and CoA to form acetyl CoA, however its significance is much larger. The most well known function of the product from this enzymatic reaction is the use of Acetyl-CoA in the role of the TCA cycle as well as in the production of fatty acid. Recent research has also shown that this enzyme is vital to the action of histone acetylation as well as gene regulation. The effect this acetylation has is far reaching in mammals. It has been shown that downregulation of the acs gene in the hippocampal region of mice results in lower levels of histone acetylation, but also impairs the long-term spatial memory of the animal. This result points to a link between cellular metabolism, gene regulation and cognitive function. This enzyme has shown to be an interesting biomarker for the presence of tumors in colorectal carcinomas. When the gene is present, the cells are able to take in acetate as a food source to convert it to Acetyl-CoA during stressed conditions. In the cases of advanced carcinoma tumors, the genes for this enzyme were down regulated and indicated a poor 5-year survival rate. Expression of the enzyme has also been linked to the development of metastatic tumor nodes, leading to a poor survival rate in patients with renal cell carcinomas.
Role in gene expression
While acetyl-CoA synthetase’s activity is usually associated with metabolic pathways, the enzyme also participates in gene expression. In yeast, acetyl-CoA synthetase delivers acetyl-CoA to histone acetyltransferases for histone acetylation. Without correct acetylation, DNA cannot condense into chromatin properly, which inevitably results in transcriptional errors.[14]
Industrial application
By taking advantage of the pathways which use acetyl-CoA as a substrate, engineered products can be obtained which have potential to be consumer products. By overexpressing the acs gene, and using acetate as a feedstock, the production of fatty acids (FAs) may be increased.[15] The use of acetate as a feed stock is uncommon, as acetate is a normal waste product of E. coli metabolism and is toxic at high levels to the organism. By adapting the E. coli to use acetate as a feedstock, these microbes were able to survive and produce their engineered products. These fatty acids could then be used as a biofuel after being separated from the media, requiring further processing (transesterification) to yield usable biodiesel fuel. Original adaptation protocol for inducing high levels of acetate uptake was innovated in 1959 as a means to induce starvation mechanisms in E. coli. [16]
Intracellular
[math]\displaystyle{ Acetate \Longrightarrow Acetyl-CoA }[/math]
[math]\displaystyle{ Acetyl-CoA \Longrightarrow FAs }[/math]
Acetyl-CoA from the breakdown of sugars in glycolysis have been sued to build fatty acids. However the difference comes in the fact that the Keasling strain is able to synthesize its own ethanol, and process (by transesterification) the fatty acid further to create stable fatty acid ethyl esters (FAEEs). Removing the need for further processing prior to obtaining a usable fuel product in Diesel engines.[17]
[math]\displaystyle{ glucose \Longrightarrow Acetyl-CoA }[/math]
Acetyl CoA used in the production of both ethanol and fatty acids
[math]\displaystyle{ Acetyl-CoA \Longrightarrow fatty acid+ethanol }[/math]
Trans-esterification
[math]\displaystyle{ fattyacid + ethanol \Longrightarrow FAEE }[/math]
Preliminary studies have been conducted where the combination of these two methods have resulted in the production of FAEEs, using acetate as the only carbon source using a combination of the methods described above.[18][unreliable source] It should be noted that the levels of production of all methods mentioned are not up to levels required for large scale applications (yet).
References
- ↑ KEGG
- ↑ 2.0 2.1 "Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2". Proceedings of the National Academy of Sciences of the United States of America 103 (27): 10224–10229. July 2006. doi:10.1073/pnas.0603968103. PMID 16788062.
- ↑ 3.0 3.1 "Transcriptional regulation of the murine acetyl-CoA synthetase 1 gene through multiple clustered binding sites for sterol regulatory element-binding proteins and a single neighboring site for Sp1". The Journal of Biological Chemistry 276 (36): 34259–69. September 2001. doi:10.1074/jbc.M103848200. PMID 11435428.
- ↑ 4.0 4.1 "Inactivation of acetyl-CoA synthase/carbon monoxide dehydrogenase by copper". Journal of the American Chemical Society 125 (31): 9316–7. August 2003. doi:10.1021/ja0352855. PMID 12889960. http://pubs.acs.org/cgi-bin/abstract.cgi/jacsat/2003/125/i31/abs/ja0352855.html.
- ↑ PDB: 1RY2; "Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP". Biochemistry 43 (6): 1425–31. February 2004. doi:10.1021/bi035911a. PMID 14769018.
- ↑ "An acetyl-CoA synthetase not encoded by the facA gene is expressed under carbon starvation in Phycomyces blakesleeanus". Research in Microbiology 156 (5–6): 663–9. Apr 7, 2005. doi:10.1016/j.resmic.2005.03.003. PMID 15921892.
- ↑ 7.0 7.1 7.2 PDB: 1PG3; "The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5'-propylphosphate and coenzyme A". Biochemistry 42 (10): 2866–73. March 2003. doi:10.1021/bi0271603. PMID 12627952.
- ↑ The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
- ↑ 9.0 9.1 "Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory" (in En). Nature 546 (7658): 381–386. June 2017. doi:10.1038/nature22405. PMID 28562591.
- ↑ "Downregulation of acetyl-CoA synthetase 2 is a metabolic hallmark of tumor progression and aggressiveness in colorectal carcinoma" (in En). Modern Pathology 30 (2): 267–277. February 2017. doi:10.1038/modpathol.2016.172. PMID 27713423.
- ↑ "Acetyl-CoA synthetase 2 enhances tumorigenesis and is indicative of a poor prognosis for patients with renal cell carcinoma". Urologic Oncology. March 2018. doi:10.1016/j.urolonc.2018.01.013. PMID 29503142.
- ↑ "Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases". Proceedings of the National Academy of Sciences of the United States of America 103 (27): 10230–10235. July 2006. doi:10.1073/pnas.0604392103. PMID 16790548.
- ↑ "Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD(+) involvement in Bacillus subtilis". Journal of Bacteriology 188 (15): 5460–8. August 2006. doi:10.1128/JB.00215-06. PMID 16855235.
- ↑ "Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription". Molecular Cell 23 (2): 207–17. July 2006. doi:10.1016/j.molcel.2006.05.040. PMID 16857587.
- ↑ Xiao, Yi; Ruan, Zhenhua; Liu, Zhiguo; Wu, Stephen G.; Varman, Arul M.; Liu, Yan; Tang, Yinjie J.. "Engineering Escherichia coli to convert acetic acid to free fatty acids". Biochemical Engineering Journal 76: 60–69. doi:10.1016/j.bej.2013.04.013.
- ↑ "The utilization of acetate-C14 by Escherichia coli grown on acetate as the sole carbon source". The Journal of Biological Chemistry 234 (8): 2118–22. August 1959. PMID 13673023. http://www.jbc.org/content/234/8/2118.
- ↑ "Microbial production of fatty-acid-derived fuels and chemicals from plant biomass" (in En). Nature 463 (7280): 559–62. January 2010. doi:10.1038/nature08721. PMID 20111002.
- ↑ "From toxic byproduct to biofuels: Adapting engineered Escherichia coli to produce fatty acid ethyl esters from acetate."". Stanford University Course: CHEMENG 185B. March 2017.