Biology:Aminoacyl tRNA synthetase
| Anticodon-binding domain of tRNA | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 leucyl-tRNA synthetase from Thermus thermophilus complexed with a post-transfer editing substrate analogue | |||||||||
| Identifiers | |||||||||
| Symbol | Anticodon_2 | ||||||||
| Pfam | PF08264 | ||||||||
| InterPro | IPR013155 | ||||||||
| SCOP2 | 1ivs / SCOPe / SUPFAM | ||||||||
| |||||||||
| DALR anticodon binding domain 1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Thermus thermophilus arginyl-trna synthetase | |||||||||
| Identifiers | |||||||||
| Symbol | DALR_1 | ||||||||
| Pfam | PF05746 | ||||||||
| Pfam clan | CL0258 | ||||||||
| InterPro | IPR008909 | ||||||||
| SCOP2 | 1bs2 / SCOPe / SUPFAM | ||||||||
| |||||||||
| DALR anticodon binding domain 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of cysteinyl-tRNA synthetase binary complex with tRNACys | |||||||||
| Identifiers | |||||||||
| Symbol | DALR_2 | ||||||||
| Pfam | PF09190 | ||||||||
| Pfam clan | CL0258 | ||||||||
| InterPro | IPR015273 | ||||||||
| |||||||||
An aminoacyl-tRNA synthetase (aaRS or ARS), also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code.
This is sometimes called "charging" or "loading" the tRNA with an amino acid. Once the tRNA is charged, a ribosome can transfer the amino acid from the tRNA onto a growing peptide, according to the genetic code. Aminoacyl tRNA therefore plays an important role in RNA translation, the expression of genes to create proteins.
Mechanism
The synthetase first binds ATP and the corresponding amino acid (or its precursor) to form an aminoacyl-adenylate, releasing inorganic pyrophosphate (PPi). The adenylate-aaRS complex then binds the appropriate tRNA molecule's D arm, and the amino acid is transferred from the aa-AMP to either the 2'- or the 3'-OH of the last tRNA nucleotide (A76) at the 3'-end.
The mechanism can be summarized in the following reaction series:
- Amino Acid + ATP → Aminoacyl-AMP + PPi
- Aminoacyl-AMP + tRNA → Aminoacyl-tRNA + AMP
Summing the reactions, the highly exergonic overall reaction is as follows:
- Amino Acid + tRNA + ATP → Aminoacyl-tRNA + AMP + PPi
Some synthetases also mediate an editing reaction to ensure high fidelity of tRNA charging. If the incorrect tRNA is added (aka. the tRNA is found to be improperly charged), the aminoacyl-tRNA bond is hydrolyzed. This can happen when two amino acids have different properties even if they have similar shapes—as is the case with valine and threonine.
The accuracy of aminoacyl-tRNA synthetase is so high that it is often paired with the word "superspecificity” when it is compared to other enzymes that are involved in metabolism. Although not all synthetases have a domain with the sole purpose of editing, they make up for it by having specific binding and activation of their affiliated amino acids. Another contribution to the accuracy of these synthetases is the ratio of concentrations of aminoacyl-tRNA synthetase and its cognate tRNA. Since tRNA synthetase improperly acylates the tRNA when the synthetase is overproduced, a limit must exist on the levels of aaRSs and tRNAs in vivo.[1][2]
Classes
There are two classes of aminoacyl tRNA synthetase, each composed of ten enzymes:[3][4]
- Class I has two highly conserved sequence motifs. It aminoacylates at the 2'-OH of a terminal adenosine nucleotide on tRNA, and it is usually monomeric or dimeric (one or two subunits, respectively).
- Class II has three highly conserved sequence motifs. It aminoacylates at the 3'-OH of a terminal adenosine on tRNA, and is usually dimeric or tetrameric (two or four subunits, respectively). Although phenylalanine-tRNA synthetase is class II, it aminoacylates at the 2'-OH.
The amino acids are attached to the hydroxyl (-OH) group of the adenosine via the carboxyl (-COOH) group.
Regardless of where the aminoacyl is initially attached to the nucleotide, the 2'-O-aminoacyl-tRNA will ultimately migrate to the 3' position via transesterification.
Bacterial aminoacyl-tRNA synthetases can be grouped as follows:[5]
| Class | Amino acids |
|---|---|
| I | Arg, Cys, Gln, Glu, Ile, Leu, Met, Trp, Tyr, Val |
| II | Ala, Asn, Asp, Gly, His, Lys, Pro, Phe, Ser, Thr |
Amino acids which use class II aaRS seem to be evolutionarily older.[6]
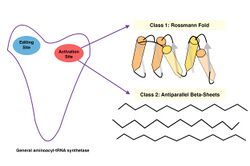

Structures
Both classes of aminoacyl-tRNA synthetases are multidomain proteins. In a typical scenario, an aaRS consists of a catalytic domain (where both the above reactions take place) and an anticodon binding domain (which interacts mostly with the anticodon region of the tRNA). Transfer-RNAs for different amino acids differ not only in their anticodon but also at other points, giving them slightly different overall configurations. The aminoacyl-tRNA synthetases recognize the correct tRNAs primarily through their overall configuration, not just through their anticodon.[7] In addition, some aaRSs have additional RNA binding domains and editing domains[8] that cleave incorrectly paired aminoacyl-tRNA molecules.
The catalytic domains of all the aaRSs of a given class are found to be homologous to one another, whereas class I and class II aaRSs are unrelated to one another. The class I aaRSs feature a cytidylyltransferase-like Rossmann fold seen in proteins like glycerol-3-phosphate cytidylyltransferase, nicotinamide nucleotide adenylyltransferase and archaeal FAD synthase, whereas the class II aaRSs have a unique fold related to biotin and lipoate ligases.
The alpha helical anticodon binding domain of arginyl-, glycyl- and cysteinyl-tRNA synthetases is known as the DALR domain after characteristic conserved amino acids.[9]
Aminoacyl-tRNA synthetases have been kinetically studied, showing that Mg2+ ions play an active catalytic role and therefore aaRs have a degree of magnesium dependence. Increasing the Mg2+ concentration leads to an increase in the equilibrium constants for the aminoacyl-tRNA synthetases’ reactions. Although this trend was seen in both class I and class II synthetases, the magnesium dependence for the two classes are very distinct. Class II synthetases have two or (more frequently) three Mg2+ ions, while class I only requires one Mg2+ ion.[10][11]
Beside their lack of overall sequence and structure similarity, class I and class II synthetases feature different ATP recognition mechanisms. While class I binds via interactions mediated by backbone hydrogen bonds, class II uses a pair of arginine residues to establish salt bridges to its ATP ligand. This oppositional implementation is manifested in two structural motifs, the Backbone Brackets and Arginine Tweezers, which are observable in all class I and class II structures, respectively. The high structural conservation of these motifs suggest that they must have been present since ancient times.[12]
Evolution
Most of the aaRSs of a given specificity are evolutionarily closer to one another than to aaRSs of another specificity. However, AsnRS and GlnRS group within AspRS and GluRS, respectively. Most of the aaRSs of a given specificity also belong to a single class. However, there are two distinct versions of the LysRS - one belonging to the class I family and the other belonging to the class II family.[citation needed]
The molecular phylogenies of aaRSs are often not consistent with accepted organismal phylogenies. That is, they violate the so-called canonical phylogenetic pattern shown by most other enzymes for the three domains of life - Archaea, Bacteria, and Eukarya. Furthermore, the phylogenies inferred for aaRSs of different amino acids often do not agree with one another. In addition, aaRS paralogs within the same species show a high degree of divergence between them. These are clear indications that horizontal transfer has occurred several times during the evolutionary history of aaRSs.[13]
A widespread belief in the evolutionary stability of this superfamily, meaning that every organism has all the aaRSs for their corresponding amino acids, is misconceived. A large-scale genomic analysis on ~2500 prokaryotic genomes showed that many of them miss one or more aaRS genes whereas many genomes have 1 or more paralogs.[14] AlaRS, GlyRS, LeuRS, IleRS and ValRS are the most evolutionarily stable members of the family. GluRS, LysRS and CysRS often have paralogs, whereas AsnRS, GlnRS, PylRS and SepRS are often absent from many genomes.
With the exception of AlaRS, it has been discovered that 19 out of the 20 human aaRSs have added at least one new domain or motif.[15] These new domains and motifs vary in function and are observed in various forms of life. A common novel function within human aaRSs is providing additional regulation of biological processes. There exists a theory that the increasing number of aaRSs that add domains is due to the continuous evolution of higher organisms with more complex and efficient building blocks and biological mechanisms. One key piece of evidence to this theory is that after a new domain is added to an aaRS, the domain becomes fully integrated. This new domain's functionality is conserved from that point on.[16]
As genetic efficiency evolved in higher organisms, 13 new domains with no obvious association with the catalytic activity of aaRSs genes have been added.
Application in biotechnology
In some of the aminoacyl tRNA synthetases, the cavity that holds the amino acid can be mutated and modified to carry unnatural amino acids synthesized in the lab, and to attach them to specific tRNAs. This expands the genetic code, beyond the twenty canonical amino acids found in nature, to include an unnatural amino acid as well. The unnatural amino acid is coded by a nonsense (TAG, TGA, TAA) triplet, a quadruplet codon, or in some cases a redundant rare codon. The organism that expresses the mutant synthetase can then be genetically programmed to incorporate the unnatural amino acid into any desired position in any protein of interest, allowing biochemists or structural biologists to probe or change the protein's function. For instance, one can start with the gene for a protein that binds a certain sequence of DNA, and, by directing an unnatural amino acid with a reactive side-chain into the binding site, create a new protein that cuts the DNA at the target-sequence, rather than binding it.
By mutating aminoacyl tRNA synthetases, chemists have expanded the genetic codes of various organisms to include lab-synthesized amino acids with all kinds of useful properties: photoreactive, metal-chelating, xenon-chelating, crosslinking, spin-resonant, fluorescent, biotinylated, and redox-active amino acids.[17] Another use is introducing amino acids bearing reactive functional groups for chemically modifying the target protein.
Certain diseases’ causation (such as neuronal pathologies, cancer, disturbed metabolic conditions, and autoimmune disorders) have been correlated to specific mutations of aminoacyl-tRNA synthetases. Charcot-Marie-Tooth (CMT) is the most frequent heritable disorder of the peripheral nervous system (a neuronal disease) and is caused by a heritable mutation in glycol-tRNA and tyrosyl-tRNA.[18] Diabetes, a metabolic disease, induces oxidative stress, which triggers a build up of mitochondrial tRNA mutations. It has also been discovered that tRNA synthetases may be partially involved in the etiology of cancer.[19] A high level of expression or modification of aaRSs has been observed within a range of cancers. A common outcome from mutations of aaRSs is a disturbance of dimer shape/formation which has a direct relationship with its function. These correlations between aaRSs and certain diseases have opened up a new door to synthesizing therapeutics.[20]
Noncatalytic domains
The novel domain additions to aaRS genes are accretive and progressive up the Tree of Life.[21][22][23] The strong evolutionary pressure for these small non-catalytic protein domains suggested their importance.[24] Findings beginning in 1999 and later revealed a previously unrecognized layer of biology: these proteins control gene expression within the cell of origin, and when released exert homeostatic and developmental control in specific human cell types, tissues and organs during adult or fetal development or both, including pathways associated with angiogenesis, inflammation, the immune response, the mechanistic target of rapamycin (mTOR) signalling, apoptosis, tumorigenesis, and interferon gamma (IFN-γ) and p53 signalling.[25][26][27][28][29][30][31][32][33]
Substrate Depletion
In 2022, it was discovered that aminoacyl-trna synthetases may incorporate alternative amino acids during shortages of their precursors. In particular, tryptophanyl-tRNA synthetase (WARS1) will incorporate phenylalanine during tryptophan depletion, essentially inducing a W>F codon reassignment.[34] Depletion of the other substrate of aminoacyl-tRNA synthetases, the cognate tRNA, may be relevant to certain diseases, e.g. Charcot–Marie–Tooth disease. It was shown that CMT-mutant glycyl-tRNA synthetase variants are still able to bind tRNA-gly but fail to release it, leading to depletion of the cellular pool of glycyl-tRNA-gly, what in turn results in stalling of the ribosome on glycine codons during mRNA translation.[35]
Clinical
Mutations in the mitochondrial enzyme have been associated with a number of genetic disorders including Leigh syndrome, West syndrome and CAGSSS (cataracts, growth hormone deficiency, sensory neuropathy, sensorineural hearing loss and skeletal dysplasia syndrome).[36]
Prediction servers
- ICAARS: B. Pawar, and GPS Raghava (2010) Prediction and classification of aminoacyl tRNA synthetases using PROSITE domains. BMC Genomics 2010, 11:507
- MARSpred: "Predicting sub-cellular localization of tRNA synthetases from their primary structures". Amino Acids 42 (5): 1703–13. May 2012. doi:10.1007/s00726-011-0872-8. PMID 21400228.
- Prokaryotic AARS database: "The complex evolutionary history of aminoacyl-tRNA synthetases". Nucleic Acids Res. 45 (3): 1059–1068. Feb 2017. doi:10.1093/nar/gkw1182. PMID 28180287.
See also
- TARS (gene)
References
- ↑ "Rules that govern tRNA identity in protein synthesis". Journal of Molecular Biology 234 (2): 257–80. November 1993. doi:10.1006/jmbi.1993.1582. PMID 8230212.
- ↑ "Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase". Science 242 (4885): 1548–51. December 1988. doi:10.1126/science.3144042. PMID 3144042. Bibcode: 1988Sci...242.1548S.
- ↑ "tRNA Synthetases". http://www.biochem.ucl.ac.uk/bsm/xtal/teach/trna/trna.html.
- ↑ Delarue, M (1995). "Aminoacyl-tRNA synthetases". Structural Biology 5 (1): 48–55. doi:10.1016/0959-440x(95)80008-o. PMID 7773747.
- ↑ Voet, Donald; Voet, Judith G. (2011). Biochemistry (4th ed.). Hoboken, NJ: Wiley. ISBN 978-0-470-57095-1.
- ↑ Trifonov, E. N (2000-12-30). "Consensus temporal order of amino acids and evolution of the triplet code" (in en). Gene. Papers presented at the Anton Dohrn Workshop 261 (1): 139–151. doi:10.1016/S0378-1119(00)00476-5. ISSN 0378-1119. PMID 11164045. https://www.sciencedirect.com/science/article/pii/S0378111900004765.
- ↑ "An operational RNA code for amino acids and possible relationship to genetic code". Proceedings of the National Academy of Sciences of the United States of America 90 (19): 8763–8. October 1993. doi:10.1073/pnas.90.19.8763. PMID 7692438. Bibcode: 1993PNAS...90.8763S.
- ↑ "Molecule of the Month: Aminoacyl-tRNA Synthetases High Fidelity". http://www.pdb.org/pdb/101/motm.do?momID=16.
- ↑ "Evolution of aminoacyl-tRNA synthetases--analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events". Genome Research 9 (8): 689–710. August 1999. doi:10.1101/gr.9.8.689. PMID 10447505.
- ↑ "Magnesium dependence of the measured equilibrium constants of aminoacyl-tRNA synthetases". Biophysical Chemistry 131 (1–3): 29–35. December 2007. doi:10.1016/j.bpc.2007.08.006. PMID 17889423.
- ↑ "Aminoacyl-tRNA synthetases in biology and disease: new evidence for structural and functional diversity in an ancient family of enzymes". RNA 3 (9): 954–60. September 1997. PMID 9292495.
- ↑ "Backbone Brackets and Arginine Tweezers delineate Class I and Class II aminoacyl tRNA synthetases". PLOS Computational Biology 14 (4): e1006101. April 2018. doi:10.1371/journal.pcbi.1006101. PMID 29659563. Bibcode: 2018PLSCB..14E6101K.
- ↑ "Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process". Microbiology and Molecular Biology Reviews 64 (1): 202–36. March 2000. doi:10.1128/MMBR.64.1.202-236.2000. PMID 10704480.
- ↑ "The complex evolutionary history of aminoacyl-tRNA synthetases". Nucleic Acids Research 45 (3): 1059–1068. February 2017. doi:10.1093/nar/gkw1182. PMID 28180287.
- ↑ "New functions of aminoacyl-tRNA synthetases beyond translation". Nature Reviews Molecular Cell Biology 11 (9): 668–74. September 2010. doi:10.1038/nrm2956. PMID 20700144.
- ↑ "Aminoacyl-tRNA synthetase complexes: beyond translation". Journal of Cell Science 117 (Pt 17): 3725–34. August 2004. doi:10.1242/jcs.01342. PMID 15286174.
- ↑ Peter G. Schultz, Expanding the genetic code
- ↑ "Crystallization and preliminary X-ray analysis of a native human tRNA synthetase whose allelic variants are associated with Charcot-Marie-Tooth disease". Acta Crystallographica Section F 62 (Pt 12): 1243–6. December 2006. doi:10.1107/S1744309106046434. PMID 17142907.
- ↑ "Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3". Proceedings of the National Academy of Sciences of the United States of America 108 (49): 19635–40. December 2011. doi:10.1073/pnas.1103922108. PMID 22106287. Bibcode: 2011PNAS..10819635K.
- ↑ "Aminoacyl tRNA synthetases and their connections to disease". Proceedings of the National Academy of Sciences of the United States of America 105 (32): 11043–9. August 2008. doi:10.1073/pnas.0802862105. PMID 18682559. Bibcode: 2008PNAS..10511043P.
- ↑ "Construction and analysis of deletions in the amino-terminal extension of glutamine tRNA synthetase of Saccharomyces cerevisiae". The Journal of Biological Chemistry 262 (22): 10807–13. August 1987. doi:10.1016/S0021-9258(18)61035-X. PMID 3301842.
- ↑ "Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs". Nature 347 (6289): 203–6. September 1990. doi:10.1038/347203a0. PMID 2203971. Bibcode: 1990Natur.347..203E.
- ↑ "Aminoacyl-tRNA synthetases". Current Opinion in Structural Biology 7 (6): 881–9. December 1997. doi:10.1016/s0959-440x(97)80161-3. PMID 9434910.
- ↑ "Human tRNA synthetase catalytic nulls with diverse functions". Science 345 (6194): 328–32. July 2014. doi:10.1126/science.1252943. PMID 25035493. Bibcode: 2014Sci...345..328L.
- ↑ "Two distinct cytokines released from a human aminoacyl-tRNA synthetase". Science 284 (5411): 147–51. April 1999. doi:10.1126/science.284.5411.147. PMID 10102815. Bibcode: 1999Sci...284..147W.
- ↑ "The evolving roles of alternative splicing". Current Opinion in Structural Biology 14 (3): 273–82. June 2004. doi:10.1016/j.sbi.2004.05.002. PMID 15193306.
- ↑ "A human aminoacyl-tRNA synthetase as a regulator of angiogenesis". Proceedings of the National Academy of Sciences of the United States of America 99 (1): 173–7. January 2002. doi:10.1073/pnas.012602099. PMID 11773626. Bibcode: 2002PNAS...99..173W.
- ↑ "VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function". The Journal of Biological Chemistry 280 (4): 2405–8. January 2005. doi:10.1074/jbc.C400431200. PMID 15579907.
- ↑ "Noncanonical activity of seryl-transfer RNA synthetase and vascular development". Trends in Cardiovascular Medicine 19 (6): 179–82. August 2009. doi:10.1016/j.tcm.2009.11.001. PMID 20211432.
- ↑ "Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality". Nature Structural & Molecular Biology 17 (1): 57–61. January 2010. doi:10.1038/nsmb.1706. PMID 20010843.
- ↑ "Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response". Proceedings of the National Academy of Sciences of the United States of America 102 (18): 6356–61. May 2005. doi:10.1073/pnas.0500226102. PMID 15851690.
- ↑ "Phosphorylation of glutamyl-prolyl tRNA synthetase by cyclin-dependent kinase 5 dictates transcript-selective translational control". Proceedings of the National Academy of Sciences of the United States of America 108 (4): 1415–20. January 2011. doi:10.1073/pnas.1011275108. PMID 21220307. Bibcode: 2011PNAS..108.1415A.
- ↑ "Essential nontranslational functions of tRNA synthetases". Nature Chemical Biology 9 (3): 145–53. March 2013. doi:10.1038/nchembio.1158. PMID 23416400.
- ↑ Pataskar, Abhijeet; Champagne, Julien; Nagel, Remco; Kenski, Juliana; Laos, Maarja; Michaux, Justine; Pak, Hui Song; Bleijerveld, Onno B. et al. (2022-03-09). "Tryptophan depletion results in tryptophan-to-phenylalanine substitutants" (in en). Nature 603 (7902): 721–727. doi:10.1038/s41586-022-04499-2. ISSN 1476-4687. PMID 35264796. Bibcode: 2022Natur.603..721P.
- ↑ Zuko, Amila; Mallik, Moushami; Thompson, Robin; Spaulding, Emily L.; Wienand, Anne R.; Been, Marije; Tadenev, Abigail L. D.; van Bakel, Nick et al. (2021-09-03). "tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase". Science 373 (6559): 1161–1166. doi:10.1126/science.abb3356. ISSN 1095-9203. PMID 34516840. Bibcode: 2021Sci...373.1161Z.
- ↑ "Expanding the clinical phenotype of IARS2-related mitochondrial disease". BMC Med Genet 19 (1): 196. 2018. doi:10.1186/s12881-018-0709-3. PMID 30419932.
External links
- Amino+Acyl-tRNA+Synthetases at the US National Library of Medicine Medical Subject Headings (MeSH)
- AARS human gene location in the UCSC Genome Browser.
- AARS human gene details in the UCSC Genome Browser.
 |
