Biology:CTP synthetase
CTP synthase is an enzyme (EC 6.3.4.2) involved in pyrimidine biosynthesis that interconverts UTP and CTP.[1][2]
Reaction mechanism
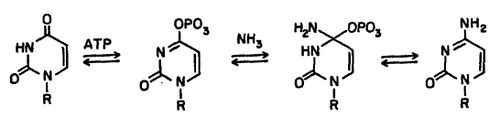
CTP (cytidine triphosphate) synthetase catalyzes the last committed step in pyrimidine nucleotide biosynthesis:[3]
ATP + UTP + glutamine → ADP + Pi + CTP + glutamate
It is the rate-limiting enzyme for the synthesis of cytosine nucleotides from both the de novo and uridine salvage pathways.[4]
The reaction proceeds by the ATP-dependent phosphorylation of UTP on the 4-oxygen atom, making the 4-carbon electrophilic and vulnerable to reaction with ammonia.[5] The source of the amino group in CTP is glutamine, which is hydrolysed in a glutamine amidotransferase domain to produce ammonia. This is then channeled through the interior of the enzyme to the synthetase domain.[6][7] Here, ammonia reacts with the intermediate 4-phosphoryl UTP.[8]
Isozymes
Two isozymes with CTP synthase activity exist in humans, encoded by the following genes:
- CTPS – CTP synthase 1
- CTPS2 – CTP synthase 2
Structure
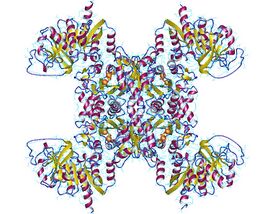
Active CTP synthase exists as a homotetrameric enzyme. At low enzyme concentrations and in the absence of ATP and UTP, CTP synthase exists as inactive monomer. As enzyme concentration increases, it polymerizes first to a dimer (such as the form shown to the left) and, in the presence of ATP and UTP, forms a tetramer.[5][9]
The enzyme contains two major domains, responsible for the aminotransferase and synthase activity, respectively. The amidotransferase domains are located away from the tetramer interfaces and are not affected by the oligomeric state. The ATP-binding site and CTP-binding site in the synthase domain are located at the tetramer interface. It is for this reason that ATP and UTP are required for tetramerization.[10]
Regulation
CTP synthase is precisely regulated by the intracellular concentrations of CTP and UTP, and both hCTPS1 and hCTPS2 have been seen to be maximally active at physiological concentrations of ATP, GTP, and glutamine.[11]
The activity of human CTPS1 isozyme has been demonstrated to be inhibited by phosphorylation.[12] One major example of this is phosphorylation of the Ser-571 residue by glycogen synthase kinase 3 (GSK3) in response to low serum conditions.[13] Additionally, Ser568 has been seen to be phosphorylated by casein kinase 1, inhibiting CTP synthase activity.[11]
CTP is also subject to various forms of allosteric regulation. GTP acts as an allosteric activator that strongly promotes the hydrolysis of glutamine, but is also inhibiting to glutamine-dependent CTP formation at high concentrations.[14] This acts to balance the relative amounts of purine and pyrimidine nucleotides. The reaction product CTP also serves as an allosteric inhibitor. The triphosphate binding site overlaps with that of UTP, but the nucleoside moiety of CTP binds in an alternative pocket opposite the binding site for UTP.[15]
CTP synthase levels have been shown to be dependent on levels of the transcription factor Myc. In turn, CTP synthase activity is required for Myc related phenotypes.[16]
The glutamine analog DON has also been seen to act as an irreversible inhibitor, and has been used as an anti-cancer agent.[17]
Filaments
CTP synthase has been reported to form filaments in several different organisms. These include bacteria (C. crescentus),[18] yeast (S. cerevisiae),[19] fruit flies (D. melanogaster)[20] and human cells.[21] These filamentous structures have been referred to as cytoplasmic rods and rings,[22] cytoophidia (from the Greek "cyto" meaning cell and "ophidium" meaning serpent, due to the structures morphology) or simply CTP synthase filaments. It has been shown that filamentation downregulates or upregulates CTP synthase activity depending on the species.[23][24][25][26][27] In Drosophila, only one of the CTP synthase isoform forms the filament.[28] Since the discovery of this novel mode of enzyme regulation in CTP synthase, multiple other enzymes have been shown to exhibit similar characteristics, suggesting that this is an important and well conserved strategy for enzymatic regulation.[29] CTP synthase remains a model enzyme for the study of filament formation.
Clinical significance
Upregulated CTP synthase activity has been widely seen in human and rodent tumors.[30] Evidence from fly models[16] and human cancer cells[31] suggests that Myc-dependent cell growth may be more susceptible to suppression of CTP-synthase activity.
Mutations in the CTP synthase have been seen to confer resistance to cytotoxic drugs such as cytosine arabinoside (ara-C) in a Chinese hamster ovary (CHO) cell model of leukemia though such mutations were not found in human patients with ara-C resistance.[32]
See also
References
- ↑ "Enzymatic amination of uridine triphosphate to cytidine triphosphate". The Journal of Biological Chemistry 222 (2): 765–775. October 1956. doi:10.1016/S0021-9258(20)89934-7. PMID 13367044.
- ↑ "The subunit structure and subunit interactions of cytidine triphosphate synthetase". The Journal of Biological Chemistry 245 (1): 80–87. January 1970. doi:10.1016/S0021-9258(18)63424-6. PMID 5411547.
- ↑ "CTP Synthetase and Related Enzymes". The Enzymes (3rd ed.). New York: Academic Press. 1974. pp. 539–59. ISBN 978-0-12-122710-4. https://books.google.com/books?id=9Jz_CZs5lFgC&pg=PA539.
- ↑ "Isoforms of human CTP synthetase". Purine and Pyrimidine Metabolism in Man X. Advances in Experimental Medicine and Biology. 486. 2000. pp. 257–261. doi:10.1007/0-306-46843-3_50. ISBN 978-0-306-46515-4.
- ↑ 5.0 5.1 "Mechanistic investigations of Escherichia coli cytidine-5'-triphosphate synthetase. Detection of an intermediate by positional isotope exchange experiments". The Journal of Biological Chemistry 260 (28): 14993–14997. December 1985. doi:10.1016/S0021-9258(18)95692-9. PMID 2933396.
- ↑ "Cytidine triphosphate synthetase. Covalent intermediates and mechanisms of action". Biochemistry 10 (18): 3365–3371. August 1971. doi:10.1021/bi00794a008. PMID 4940761.
- ↑ "Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets". Biochemistry 43 (21): 6447–6463. June 2004. doi:10.1021/bi0496945. PMID 15157079.
- ↑ "Investigation of the mechanism of CTP synthetase using rapid quench and isotope partitioning methods". Biochemistry 28 (21): 8454–8459. October 1989. doi:10.1021/bi00447a027. PMID 2532543.
- ↑ "CTP synthetase from Escherichia coli: an improved purification procedure and characterization of hysteretic and enzyme concentration effects on kinetic properties". Biochemistry 22 (13): 3285–3292. June 1983. doi:10.1021/bi00282a038. PMID 6349684.
- ↑ "Structure of the dimeric form of CTP synthase from Sulfolobus solfataricus". Acta Crystallographica. Section F, Structural Biology and Crystallization Communications 67 (Pt 2): 201–208. February 2011. doi:10.1107/S1744309110052334. PMID 21301086.
- ↑ 11.0 11.1 "Regulation of human cytidine triphosphate synthetase 2 by phosphorylation". The Journal of Biological Chemistry 285 (44): 33727–33736. October 2010. doi:10.1074/jbc.M110.178566. PMID 20739275.
- ↑ "Phospholipid synthesis in yeast: regulation by phosphorylation". Biochemistry and Cell Biology 82 (1): 62–70. February 2004. doi:10.1139/o03-064. PMID 15052328.
- ↑ "Regulation of human cytidine triphosphate synthetase 1 by glycogen synthase kinase 3". The Journal of Biological Chemistry 282 (40): 29493–29503. October 2007. doi:10.1074/jbc.M703948200. PMID 17681942.
- ↑ "Structural requirements for the activation of Escherichia coli CTP synthase by the allosteric effector GTP are stringent, but requirements for inhibition are lax". The Journal of Biological Chemistry 283 (4): 2010–2020. January 2008. doi:10.1074/jbc.M707803200. PMID 18003612.
- ↑ "Mechanisms of product feedback regulation and drug resistance in cytidine triphosphate synthetases from the structure of a CTP-inhibited complex". Biochemistry 44 (41): 13491–13499. October 2005. doi:10.1021/bi051282o. PMID 16216072.
- ↑ 16.0 16.1 "The Interplay between Myc and CTP Synthase in Drosophila". PLOS Genetics 12 (2): e1005867. February 2016. doi:10.1371/journal.pgen.1005867. PMID 26889675.
- ↑ "Metabolism and action of amino acid analog anti-cancer agents". Pharmacology & Therapeutics 46 (2): 243–271. 1990. doi:10.1016/0163-7258(90)90094-I. PMID 2108451. https://zenodo.org/record/1258343.
- ↑ "The metabolic enzyme CTP synthase forms cytoskeletal filaments". Nature Cell Biology 12 (8): 739–746. August 2010. doi:10.1038/ncb2087. PMID 20639870.
- ↑ "Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster". The Journal of Cell Biology 190 (4): 541–551. August 2010. doi:10.1083/jcb.201003001. PMID 20713603.
- ↑ "Intracellular compartmentation of CTP synthase in Drosophila". Journal of Genetics and Genomics = Yi Chuan Xue Bao 37 (5): 281–296. May 2010. doi:10.1016/S1673-8527(09)60046-1. PMID 20513629. https://ora.ox.ac.uk/objects/uuid:c0636dbd-1c5f-4663-ae54-1589f8582b09.
- ↑ "Glutamine analogs promote cytoophidium assembly in human and Drosophila cells". Journal of Genetics and Genomics = Yi Chuan Xue Bao 38 (9): 391–402. September 2011. doi:10.1016/j.jgg.2011.08.004. PMID 21930098.
- ↑ "Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells". PLOS ONE 6 (12): e29690. 2011. doi:10.1371/journal.pone.0029690. PMID 22220215. Bibcode: 2011PLoSO...629690C.
- ↑ "Human CTP synthase filament structure reveals the active enzyme conformation". Nature Structural & Molecular Biology 24 (6): 507–514. June 2017. doi:10.1038/nsmb.3407. PMID 28459447.
- ↑ "Large-scale filament formation inhibits the activity of CTP synthetase". eLife 3: e03638. July 2014. doi:10.7554/eLife.03638. PMID 25030911.
- ↑ "Nucleotide synthesis is regulated by cytoophidium formation during neurodevelopment and adaptive metabolism". Biology Open 3 (11): 1045–1056. October 2014. doi:10.1242/bio.201410165. PMID 25326513.
- ↑ "Common regulatory control of CTP synthase enzyme activity and filament formation". Molecular Biology of the Cell 25 (15): 2282–2290. August 2014. doi:10.1091/mbc.E14-04-0912. PMID 24920825.
- ↑ "Self-assembling enzymes and the origins of the cytoskeleton". Current Opinion in Microbiology 14 (6): 704–711. December 2011. doi:10.1016/j.mib.2011.09.015. PMID 22014508.
- ↑ "Only one isoform of Drosophila melanogaster CTP synthase forms the cytoophidium". PLOS Genetics 9 (2): e1003256. February 2013. doi:10.1371/journal.pgen.1003256. PMID 23459760.
- ↑ "Metabolic regulation via enzyme filamentation". Critical Reviews in Biochemistry and Molecular Biology 51 (4): 282–293. 2015. doi:10.3109/10409238.2016.1172555. PMID 27098510.
- ↑ "Increased cytidine 5'-triphosphate synthetase activity in rat and human tumors". Cancer Research 40 (11): 3921–3927. November 1980. PMID 7471043. http://cancerres.aacrjournals.org/cgi/pmidlookup?view=long&pmid=7471043.
- ↑ "Combined Inactivation of CTPS1 and ATR Is Synthetically Lethal to MYC-Overexpressing Cancer Cells". Cancer Research 82 (6): 1013–1024. March 2022. doi:10.1158/0008-5472.can-21-1707. PMID 35022212.
- ↑ "Resistance to cytosine arabinoside in acute leukemia: the significance of mutations in CTP synthetase". Leukemia 8 (2): 264–265. February 1994. PMID 8309250.
Further reading
- "Immunology: When lymphocytes run out of steam". Nature 510 (7504): 222–223. June 2014. doi:10.1038/nature13346. PMID 24870231. Bibcode: 2014Natur.510..222V.
External links
- CTP+synthetase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |