Biology:C1orf27
Uncharacterized protein Chromosome 1 Open Reading Frame 27 is a protein in humans, encoded by the C1orf27 gene. It is accession number NM_017847.[1] This is a membrane protein that is 3926 base pairs long with the most extensive string of amino acids being 454aa long. C1orf27 exhibits cytoplasmic expression in epidermal tissues.[2] Predicted associated biological processes of the gene include cell fate specification and developmental properties.[citation needed]
Gene
Locus
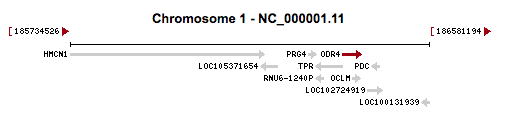
This gene is located on chromosome 1 at 1q31.1.[3] It is encoded on the plus strand of DNA spanning from 186,344,406 to 186,390,514.
mRNA
Alternative splicing
There appear to be four isoforms due to splicing.[4] Two of those are truncated on the 3' end of the protein from 266aa and 396aa. Additional location of alternative splice sites are from 79aa to 102aa and 246aa to 260aa.
Protein
General properties
The primary encoded protein of C1orf27 consists of 454 amino acid residues and is 3926 base pairs long.[1] It consists of 14 total exons. The predicted molecular weight of the primary, unmodified protein is approximately 51.1 kdal.
Aliases
As with many other genes, there are some common aliases found with this gene.[citation needed] Those aliases are Lymphocyte-Activation Gene-1 (LAG1) Interacting Protein, Transparent Testa Glabra 1 (TTG1), and Odorant Response Abnormal 4 (ODR4). The most common alias for C1orf27 is ODR4, and this is what most readily appears when searching the gene.
Composition
Computational analysis revealed the most abundant amino acid to be leucine at 10.1% of the total protein.[5] The second most abundant was serine which contributes to 8.6% of the total protein. Glutamic acid was third most abundant and contributes to 7.7% of the protein. This analysis also revealed that the protein appears to be deficient in tryptophan as it only contributes to 1.1% of the protein.[5] Based on the distribution of other amino acid types, there were five high scoring hydrophobic segments. There were also two transmembrane domains located at 82-98aa and 432-449aa.
Post-translational modifications
| Interactor | Number of Predicted Sites | Function |
|---|---|---|
| N-myristoylation | 8 | Key components of signaling pathways, and typically promotes membrane binding essential for protein localization and/ or biological function[6] |
| N-glycosylation | 4 | Increase protein stability by decreasing protein dynamics.[7] |
| Protein Kinase C Phosphorylation | 7 | Enzymatic activity regulation.[8] |
| Casein Kinase II Phosphorylation | 7 | Epidermal growth factor role.[9] |
| Tyrosine Kinase Phosphorylation | 2 | Alterations to the structural conformation.[10] |
| cAMP and cGMP Dependent Phosphorylation | 2 | Coordination of the active site conformation and enzymatic activity. |
C1orf27 is predicted to undergo multiple post translational modifications such as glycosylation, myristoylation, and phosphorylation.[11]
Interactions
There were eight interactions identified by Mentha.[12] The first one was UFSP2 which hydrolyzes the peptide bond at the C-term gly of UFM1, a ubiquitin-like modifier protein bound to a number of target proteins. The second one was HSCB which acts as a co-chaperone in iron-sulfur cluster assembly in mitochondria. The third was GRB2 which is an adapter protein that provides a critical link between cell surface growth factor receptors and the Ras signaling pathway. The fourth was CYLD which is a protease that cleaves Lys-63-linked polyubiquitin chains, controls regulation of cell survival, proliferation, and differentiation, and is required for normal cell cycle progress. The fifth was ATM which activates checkpoint signaling upon double strand breaks, apoptosis, and genotoxic stress. The sixth was FAM177A1, the function of which is unknown. The last two were THID2 and Q81kP6 which are both in bacillus anthracis.
Subcellular localization
The c1orf27 protein is likely cytoplasmic.[13] This was found with 55.5 reliability. The K-NN prediction was k=9/23 and the protein was found to be 55.6% cytoplasmic, 11.1% mitochondrial, 11.1% vacuolar, 11.1% cytoskeletal, and 11.1% golgi.
Structure
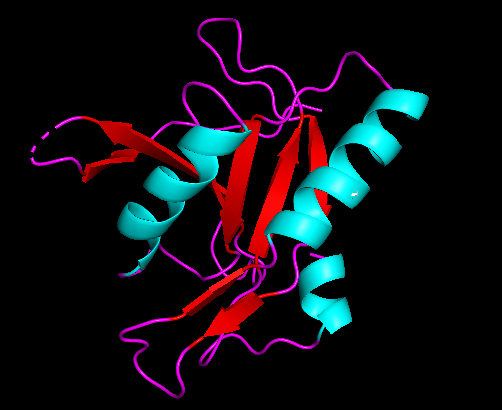
Alpha helices predicted in the c1orf27 protein are colored blue in the above picture. Beta sheets are pictured by the red arrows. Random coils are the purple strands between structures.
Expression

Overall, expression of c1orf27 seems to be ubiquitous.[16] Highest expression body sites (>50 TPM) were bladder, bone marrow, kidney, liver, pancreas, parathyroid, and vascular. Highest expression health sites (>50 TPM) were adrenal tumors, cervical tumors, and liver tumors. While both of these observations had relatively high TPM scores, there was still relatively low occurrence. This validates the assumption that expression is ubiquitous. There was moderate expression (>25 TPM) in the human fetus, and expression increased with age.[16] Expression was completely absent in the ears, esophagus, lymph, nerve, salivary glands, thyroid, tonsils, and umbilical cord. There was no expression in bladder carcinoma despite expression being elevated in the bladder itself. There was high expression in endothelial cells and neuronal cells but was undetectable in glial cells and neuropil cells. Expression was also localized to the nucleoplasm and plasma membrane in humans but is localized to the cytosol in mice.
Homology
Paralogs
There were no paralogs of C1orf27 identified in the human genome.[4]
Orthologs
There were orthologs identified in most animals for which there were complete genome data.[4] The most distant, yet still relevant, orthologs identified were invertebrates from phylum Cnidaria.
Molecular Evolution
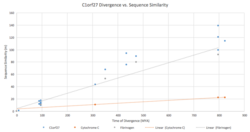
The m value, or number of corrected amino acid changes per 100 residues, for the C1orf27 gene was graphed against the species divergence in millions of years. When compared to divergence graphs of fibrinogen and cytochrome C, it was determined that this gene closely resembles the evolutionary pattern observed in fibrinogen, suggesting a more rapid rate of evolution. M values for C1orf27 were calculated using the percentage of identity, when compared to humans, observed in the mRNA sequences of the orthologs using the formula derived from the Molecular Clock Hypothesis.
References
- ↑ 1.0 1.1 "Homo sapiens odr-4 GPCR localization factor homolog (ODR4), transcript - Nucleotide - NCBI". https://www.ncbi.nlm.nih.gov/nuccore/NM_017847.5.
- ↑ "Tissue expression of C1orf27 - Summary - The Human Protein Atlas". https://www.proteinatlas.org/ENSG00000157181-C1orf27/tissue.
- ↑ 3.0 3.1 "ODR4 odr-4 GPCR localization factor homolog [Homo sapiens (human) - Gene - NCBI"]. https://www.ncbi.nlm.nih.gov/gene/54953.
- ↑ 4.0 4.1 4.2 "Protein BLAST: search protein databases using a protein query" (in en). https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome.
- ↑ 5.0 5.1 EMBL-EBI. "SAPS < Sequence Statistics < EMBL-EBI" (in en). https://www.ebi.ac.uk/Tools/seqstats/saps/.
- ↑ "Protein myristoylation in health and disease". Journal of Chemical Biology 3 (1): 19–35. March 2010. doi:10.1007/s12154-009-0032-8. PMID 19898886.
- ↑ "Proteomics/Post-translational Modification/Glycosylation - Wikibooks, open books for an open world" (in en). https://en.wikibooks.org/wiki/Proteomics/Post-translational_Modification/Glycosylation.
- ↑ "Posttranslational modifications on protein kinase c isozymes. Effects of epinephrine and phorbol esters". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1783 (5): 695–712. May 2008. doi:10.1016/j.bbamcr.2007.07.011. PMID 18295358.
- ↑ "Casein kinase II phosphorylates the fragile X mental retardation protein and modulates its biological properties". Molecular and Cellular Biology 22 (24): 8438–47. December 2002. doi:10.1128/MCB.22.24.8438-8447.2002. PMID 12446764.
- ↑ "Emerging roles of post-translational modifications in signal transduction and angiogenesis". Proteomics 15 (2–3): 300–9. January 2015. doi:10.1002/pmic.201400183. PMID 25161153.
- ↑ "ExPASy - PROSITE" (in en-US). https://prosite.expasy.org/.
- ↑ "mentha: the interactome browser". https://mentha.uniroma2.it/.
- ↑ "PSORT II Prediction". https://psort.hgc.jp/form2.html.
- ↑ Kelley, Lawrence. "PHYRE2 Protein Fold Recognition Server". http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index.
- ↑ "GDS1402 / NM_017847.1_PROBE1". https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS1402:NM_017847.1_PROBE1.
- ↑ 16.0 16.1 Group, Schuler. "EST Profile - Hs.371210". https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.371210.
 |