Biology:Channelrhodopsin
Channelrhodopsins are a subfamily of retinylidene proteins (rhodopsins) that function as light-gated ion channels.[1] They serve as sensory photoreceptors in unicellular green algae, controlling phototaxis: movement in response to light.[2] Expressed in cells of other organisms, they enable light to control electrical excitability, intracellular acidity, calcium influx, and other cellular processes (see optogenetics). Channelrhodopsin-1 (ChR1) and Channelrhodopsin-2 (ChR2) from the model organism Chlamydomonas reinhardtii are the first discovered channelrhodopsins. Variants that are sensitive to different colors of light or selective for specific ions (ACRs, KCRs) have been cloned from other species of algae and protists.
History
Phototaxis and photoorientation of microalgae have been studied over more than a hundred years in many laboratories worldwide. In 1980, Ken Foster developed the first consistent theory about the functionality of algal eyes.[3] He also analyzed published action spectra and complemented blind cells with retinal and retinal analogues, which led to the conclusion that the photoreceptor for motility responses in Chlorophyceae is rhodopsin.[4]
Photocurrents of the Chlorophyceae Heamatococcus pluvialis and Chlamydomonas reinhardtii were studied over many years in the groups of Oleg Sineshchekov and Peter Hegemann.[5][6] Based on action spectroscopy and simultaneous recordings of photocurrents and flagellar beating, it was determined that the photoreceptor currents and subsequent flagellar movements are mediated by rhodopsin and control phototaxis and photophobic responses. The extremely fast rise of the photoreceptor current after a brief light flash led to the conclusion that the rhodopsin and the channel are intimately linked in a protein complex, or even within one single protein.[7][8]
The name "channelrhodopsin" was coined to highlight this unusual property, and the sequences were renamed accordingly. The nucleotide sequences of the rhodopsins now called channelrhodopsins ChR1 and ChR2 were finally uncovered in a large-scale EST sequencing project in C. reinhardtii. Independent submission of the same sequences to GenBank by three research groups generated confusion about their naming: The names cop-3 and cop-4 were used for initial submission by Hegemann's group;[9] csoA and csoB by Spudich's group;[2] and acop-1 and acop-2 by Takahashi's group.[10] Both sequences were found to function as single-component light-activated cation channels in a Xenopus oocytes and human kidney cells (HEK).[1][11]
Their roles in generation of photoreceptor currents in algal cells were characterized by Oleg Sineshchekov, Kwang-Hwan Jung and John Spudich,[2] and Peter Berthold and Peter Hegemann.[12]
Structure
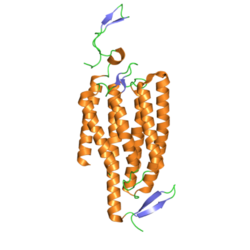
In terms of structure, channelrhodopsins are retinylidene proteins. They are seven-transmembrane proteins like rhodopsin, and contain the light-isomerizable chromophore all-trans-retinal (an aldehyde derivative of vitamin A). The retinal chromophore is covalently linked to the rest of the protein through a protonated Schiff base. Whereas most 7-transmembrane proteins are G protein-coupled receptors that open other ion channels indirectly via second messengers (i.e., they are metabotropic), channelrhodopsins directly form ion channels (i.e., they are ionotropic).[11] This makes cellular depolarization extremely fast, robust, and useful for bioengineering and neuroscience applications, including photostimulation.
Function
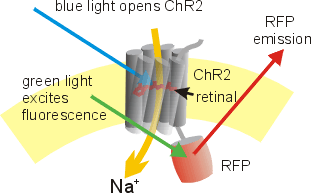
The natural ("wild-type") ChR2 absorbs blue light with an absorption and action spectrum maximum at 480 nm.[14] When the all-trans-retinal complex absorbs a photon, it induces a conformational change from all-trans to 13-cis-retinal. This change introduces a further one in the transmembrane protein, opening the pore to at least 6 Å. Within milliseconds, the retinal relaxes back to the all-trans form, closing the pore and stopping the flow of ions.[11] Most natural channelrhodopsins are nonspecific cation channels (CCRs), conducting H+, Na+, K+, and Ca2+ ions. Recently discovered anion-conducting channelrhodopsins (ACRs)[15] and potassium-selective channelrhodpsins (HcKCR1, HcKCR2) [16] were structurally analyzed to understand their ion selectivity.[17][18] ACRs and KCRs have been used to inhibit neuronal activity.
Development as a molecular tool
In 2005, three groups sequentially established ChR2 as a tool for genetically targeted optical remote control (optogenetics) of neurons, neural circuits and behavior.
At first, Karl Deisseroth's lab demonstrated that ChR2 could be deployed to control mammalian neurons in vitro, achieving temporal precision on the order of milliseconds (both in terms of delay to spiking and in terms of temporal jitter).[19] Because all opsins require retinal as the light-sensing co-factor and it was unclear whether central mammalian nerve cells would contain sufficient retinal levels, but they do. It also showed, despite the small single-channel conductance, sufficient potency to drive mammalian neurons above action potential threshold. From this, channelrhodopsin became the first optogenetic tool, with which neural activity could be controlled with the temporal precision at which neurons operate (milliseconds). A second study was published later confirming the ability of ChR2 to control the activity of vertebrate neurons, at this time in the chick spinal cord.[20] This study was the first wherein ChR2 was expressed alongside an optical silencer, vertebrate rhodopsin-4 in this case, demonstrating for the first time that excitable cells could be activated and silenced using these two tools simultaneously, illuminating the tissue at different wavelengths.
It was demonstrated that ChR2, if expressed in specific neurons or muscle cells, can evoke predictable behaviors, i.e. can control the nervous system of an intact animal, in this case the invertebrate C. elegans.[21] This was the first using ChR2 to steer the behavior of an animal in an optogenetic experiment, rendering a genetically specified cell type subject to optical remote control. Although both aspects had been illustrated earlier that year by the group of Gero Miesenböck, deploying the indirectly light-gated ion channel P2X2,[22] it was henceforth microbial opsins like channelrhodopsin that dominated the field of genetically targeted remote control of excitable cells, due to the power, speed, targetability, ease of use, and temporal precision of direct optical activation, not requiring any external chemical compound such as caged ligands.[23]
To overcome its principal downsides — the small single-channel conductance (especially in steady-state), the limitation to one optimal excitation wavelength (~470 nm, blue) as well as the relatively long recovery time, not permitting controlled firing of neurons above 20–40 Hz — ChR2 has been optimized using genetic engineering. A point mutation H134R (exchanging the amino acid Histidine in position 134 of the native protein for an Arginine) resulted in increased steady-state conductance, as described in a 2005 paper that also established ChR2 as an optogenetic tool in C. elegans.[21] In 2009, Roger Tsien's lab optimized ChR2 for further increases in steady-state conductance and dramatically reduced desensitization by creating chimeras of ChR1 and ChR2 and mutating specific amino acids, yielding ChEF and ChIEF, which allowed the driving of trains of action potentials up to 100 Hz.[24][25] In 2010, the groups of Hegemann and Deisseroth introduced an E123T mutation into native ChR2, yielding ChETA, which has faster on- and off-kinetics, permitting the control of individual action potentials at frequencies up to 200 Hz (in appropriate cell types).[26][24]
The groups of Hegemann and Deisseroth also discovered that the introduction of the point mutation C128S makes the resulting ChR2-derivative a step-function tool: Once "switched on" by blue light, ChR2(C128S) stays in the open state until it is switched off by yellow light – a modification that deteriorates temporal precision, but increases light sensitivity by two orders of magnitude.[27] They also discovered and characterized VChR1 in the multicellular algae Volvox carteri. VChR1 produces only tiny photocurrents, but with an absorption spectrum that is red-shifted relative to ChR2.[28] Using parts of the ChR1 sequence, photocurrent amplitude was later improved to allow excitation of two neuronal populations at two distinct wavelengths.[29]
Deisseroth's group has pioneered many applications in live animals such as genetically targeted remote control in rodents in vivo,[30] the optogenetic induction of learning in rodents,[31] the experimental treatment of Parkinson's disease in rats,[32][33] and the combination with fMRI (opto-fMRI).[34] Other labs have pioneered the combination of ChR2 stimulation with calcium imaging for all-optical experiments,[35] mapping of long-range[36] and local[37] neural circuits, ChR2 expression from a transgenic locus – directly[38] or in the Cre-lox conditional paradigm[37] – as well as the two-photon excitation of ChR2, permitting the activation of individual cells.[39][40][41]
In March 2013, the Brain Prize (Grete Lundbeck European Brain Research Prize) was jointly awarded to Bamberg, Boyden, Deisseroth, Hegemann, Miesenböck, and Nagel for "their invention and refinement of optogenetics".[42] The same year, Hegemann and Nagel received the Louis-Jeantet Prize for Medicine for "the discovery of channelrhodopsin". In 2015, Boyden and Deisseroth received the Breakthrough Prize in Life Sciences and in 2020, Miesenböck, Hegemann and Nagel received the Shaw prize in Life Science and Medicine for the development of optogenetics.
Designer-channelrhodopsins
Channelrhodopsins are key tools in optogenetics. The C-terminal end of Channelrhodopsin-2 extends into the intracellular space and can be replaced by fluorescent proteins without affecting channel function. This kind of fusion construct can be useful to visualize the morphology of ChR2 expressing cells, i.e. simultaneously indicate which cells are tagged with FP and allow the activity to be controlled by the channelrhodopsin.[19][35] Point mutations close to the retinal binding pocket have been shown to affect the biophysical properties of the channelrhodopsin, resulting in a variety of different tools.
Kinetics
Closing of the channel after optical activation can be substantially delayed by mutating the protein residues C128 or D156. This modification results in super-sensitive channelrhodopsins that can be opened by a blue light pulse and closed by a green or yellow light pulse (Step-function opsins).[27][43][29] Mutating the E123 residue accelerates channel kinetics (ChETA), and the resulting ChR2 mutants have been used to spike neurons at up to 200 Hz.[26] In general, channelrhodopsins with slow kinetics are more light-sensitive on the population level, as open channels accumulate over time even at low light levels.
Photocurrent amplitude
H134R and T159C mutants display increased photocurrents, and a combination of T159 and E123 (ET/TC) has slightly larger photocurrents and slightly faster kinetics than wild-type ChR2.[44] ChIEF, a chimera and point mutant of ChR1 and ChR2, demonstrates large photocurrents, little desensitization and kinetics similar to wild-type ChR2.[24] Variants with extended open time (ChR2-XXL) produce extremely large photocurrents and are very light sensitive on the population level.[45]
Wavelength
Chimeric channelrhodopsins have been developed by combining transmembrane helices from ChR1 and VChR1, leading to the development of ChRs with red spectral shifts (such as C1V1 and ReaChR).[29][46] ReaChR has improved membrane trafficking and strong expression in mammalian cells, and has been used for minimally invasive, transcranial activation of brainstem motoneurons. Searches for homologous sequences in other organisms has yielded spectrally improved and stronger red-shifted channelrhodpsins (Chrimson).[47] In combination with ChR2, these yellow/red light-sensitive channelrhodopsins allow controlling two populations of neurons independently with light pulses of different colors.[48][49]
A blue-shifted channelrhodopsin has been discovered in the alga Scherffelia dubia. After some engineering to improve membrane trafficking and speed, the resulting tool (CheRiff) produced large photocurrents at 460 nm excitation.[50] It has been combined with the Genetically Encoded Calcium Indicator jRCaMP1b [51] in an all-optical system called the OptoCaMP.[52]
Ion selectivity
Most channelrhodopsins are unspecific cation channels. When expressed in neurons, they conduct mostly Na+ ions and are therefore depolarizing (excitatory). Variants with moderate to high calcium permeability have been engineered (CatCh, CapChRs).[53][54] K+-specific channelrhodopsins (KCRs, WiChR) were recently discovered in various protists.[55][56] When expressed in neurons, potassium-selective channelrhodopsins hyperpolarize the membrane upon illumination, preventing spike generation (inhibitory).
Mutating E90 to the positively charged amino acid arginine turns channelrhodopsin from an unspecific cation channel into a chloride-conducting channel (ChloC).[57] The selectivity for Cl- was further improved by replacing negatively charged residues in the channel pore, making the reversal potential more negative.[58][59] Anion-conducting channelrhodopsins (iChloC, iC++, GtACR) inhibit neuronal spiking in cell culture and in intact animals when illuminated with blue light.
Applications
Channelrhodopsins can be readily expressed in excitable cells such as neurons using a variety of transfection techniques (viral transfection, electroporation, gene gun) or transgenic animals. The light-absorbing pigment retinal is present in most cells (of vertebrates) as Vitamin A, making it possible to photostimulate neurons without adding any chemical compounds. Before the discovery of channelrhodopsins, neuroscientists were limited to recording the activity of neurons in the brain and correlate this activity with behavior. This is not sufficient to prove that the recorded neural activity actually caused that behavior. Controlling networks of genetically modified cells with light, an emerging field known as Optogenetics., allows researchers now to explore the causal link between activity in a specific group of neurons and mental events, e.g. decision making. Optical control of behavior has been demonstrated in nematodes, fruit flies, zebrafish, and mice.[60][61] Recently, chloride-conducting channelrhodopsins have been engineered and were also found in nature.[15][57] These tools can be used to silence neurons in cell culture and in live animals by shunting inhibition.[58][59]
Using multiple colors of light expands the possibilities of optogenetic experiments. The blue-light sensitive ChR2 and the yellow light-activated chloride pump halorhodopsin together enable multiple-color optical activation and silencing of neural activity.[62][63] Another interesting pair is the blue-light sensitive chloride channel GtACR2[64] and the red-light sensitive cation channel Chrimson[65] which have been combined in a single protein (BiPOLES) for bidirectional control of membrane potential.[66]
Using fluorescently labeled ChR2, light-stimulated axons and synapses can be identified.[35] This is useful to study the molecular events during the induction of synaptic plasticity.[67][68] Transfected cultured neuronal networks can be stimulated to perform some desired behaviors for applications in robotics and control.[69] ChR2 has also been used to map long-range connections from one side of the brain to the other, and to map the spatial location of inputs on the dendritic tree of individual neurons.[36][70]
In 2006, it was reported that transfection with Channelrhodopsin could restore eyesight to blind mice.[71]
Neurons can tolerate ChR expression for a long time, and several laboratories are testing optogenetic stimulation to solve medical needs. In blind mice, visual function can be partially restored by expressing ChR2 in inner retinal cells.[72][73] In 2021, the red-light sensitive ChR ChrimsonR was virally delivered to the eyes of a human patient suffering from retinal degeneration (retinitis pigmentosa), leading to partial recovery of his vision.[74][75] Optical cochlear implants have been shown to work well in animal experiments and are currently undergoing clinical trials.[76][77][78] In the future, ChRs may find even more medical applications, e.g. for deep-brain stimulation of Parkinson patients or to control certain forms of epilepsy.
References
- ↑ 1.0 1.1 "Channelrhodopsin-1: a light-gated proton channel in green algae". Science 296 (5577): 2395–2398. June 2002. doi:10.1126/science.1072068. PMID 12089443. Bibcode: 2002Sci...296.2395N.
- ↑ 2.0 2.1 2.2 "Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii". Proceedings of the National Academy of Sciences of the United States of America 99 (13): 8689–8694. June 2002. doi:10.1073/pnas.122243399. PMID 12060707.
- ↑ "Light Antennas in phototactic algae". Microbiological Reviews 44 (4): 572–630. December 1980. doi:10.1128/mr.44.4.572-630.1980. PMID 7010112.
- ↑ "A rhodopsin is the functional photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas". Nature 311 (5988): 756–759. October 1984. doi:10.1038/311756a0. PMID 6493336. Bibcode: 1984Natur.311..756F.
- ↑ "Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis". Nature 271 (5644): 476–478. February 1978. doi:10.1038/271476a0. PMID 628427. Bibcode: 1978Natur.271..476L.
- ↑ "Rhodopsin-regulated calcium currents in Chlamydomonas.". Nature 351 (6326): 489–491. June 1991. doi:10.1038/351489a0. Bibcode: 1991Natur.351..489H.
- ↑ "The nature of rhodopsin-triggered photocurrents in Chlamydomonas. I. Kinetics and influence of divalent ions". Biophysical Journal 70 (2): 924–931. February 1996. doi:10.1016/S0006-3495(96)79635-2. PMID 8789109. Bibcode: 1996BpJ....70..924H.
- ↑ "Two light-activated conductances in the eye of the green alga Volvox carteri". Biophysical Journal 76 (3): 1668–1678. March 1999. doi:10.1016/S0006-3495(99)77326-1. PMID 10049347. Bibcode: 1999BpJ....76.1668B.
- ↑ Kateriya, S. Fuhrmann, M. Hegemann, P.: Direct Submission: Chlamydomonas reinhardtii retinal binding protein (cop4) gene; GenBank accession number AF461397
- ↑ "Archaeal-type rhodopsins in Chlamydomonas: model structure and intracellular localization". Biochemical and Biophysical Research Communications 301 (3): 711–717. February 2003. doi:10.1016/S0006-291X(02)03079-6. PMID 12565839.
- ↑ 11.0 11.1 11.2 "Channelrhodopsin-2, a directly light-gated cation-selective membrane channel". Proceedings of the National Academy of Sciences of the United States of America 100 (24): 13940–13945. November 2003. doi:10.1073/pnas.1936192100. PMID 14615590. Bibcode: 2003PNAS..10013940N.
- ↑ "Channelrhodopsin-1 initiates phototaxis and photophobic responses in chlamydomonas by immediate light-induced depolarization". The Plant Cell 20 (6): 1665–1677. June 2008. doi:10.1105/tpc.108.057919. PMID 18552201.
- ↑ "Crystal structure of the channelrhodopsin light-gated cation channel". Nature 482 (7385): 369–374. January 2012. doi:10.1038/nature10870. PMID 22266941. Bibcode: 2012Natur.482..369K.
- ↑ "Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function". Journal of Molecular Biology 375 (3): 686–694. January 2008. doi:10.1016/j.jmb.2007.10.072. PMID 18037436.
- ↑ 15.0 15.1 "NEUROSCIENCE. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics". Science 349 (6248): 647–650. August 2015. doi:10.1126/science.aaa7484. PMID 26113638. Bibcode: 2015Sci...349..647G.
- ↑ "Kalium channelrhodopsins are natural light-gated potassium channels that mediate optogenetic inhibition". Nature Neuroscience 25 (7): 967–974. July 2022. doi:10.1038/s41593-022-01094-6. PMID 35726059.
- ↑ "Crystal structure of the natural anion-conducting channelrhodopsin GtACR1". Nature 561 (7723): 343–348. September 2018. doi:10.1038/s41586-018-0511-6. PMID 30158696. Bibcode: 2018Natur.561..343K.
- ↑ "Structural basis for ion selectivity in potassium-selective channelrhodopsins" (in en). bioRxiv. 2022-10-31. doi:10.1101/2022.10.30.514430. https://www.biorxiv.org/content/biorxiv/early/2022/10/31/2022.10.30.514430.full.pdf.
- ↑ 19.0 19.1 "Millisecond-timescale, genetically targeted optical control of neural activity". Nature Neuroscience 8 (9): 1263–1268. September 2005. doi:10.1038/nn1525. PMID 16116447.
- ↑ "Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin". Proceedings of the National Academy of Sciences of the United States of America 102 (49): 17816–17821. December 2005. doi:10.1073/pnas.0509030102. PMID 16306259. Bibcode: 2005PNAS..10217816L.
- ↑ 21.0 21.1 "Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses". Current Biology 15 (24): 2279–2284. December 2005. doi:10.1016/j.cub.2005.11.032. PMID 16360690.
- ↑ "Remote control of behavior through genetically targeted photostimulation of neurons". Cell 121 (1): 141–152. April 2005. doi:10.1016/j.cell.2005.02.004. PMID 15820685.
- ↑ "Channelrhodopsin-2 and optical control of excitable cells". Nature Methods 3 (10): 785–792. October 2006. doi:10.1038/nmeth936. PMID 16990810.
- ↑ 24.0 24.1 24.2 "A user's guide to channelrhodopsin variants: features, limitations and future developments". Experimental Physiology 96 (1): 19–25. January 2011. doi:10.1113/expphysiol.2009.051961. PMID 20621963.
- ↑ "Characterization of engineered channelrhodopsin variants with improved properties and kinetics". Biophysical Journal 96 (5): 1803–1814. March 2009. doi:10.1016/j.bpj.2008.11.034. PMID 19254539. Bibcode: 2009BpJ....96.1803L.
- ↑ 26.0 26.1 "Ultrafast optogenetic control". Nature Neuroscience 13 (3): 387–392. March 2010. doi:10.1038/nn.2495. PMID 20081849.
- ↑ 27.0 27.1 "Bi-stable neural state switches". Nature Neuroscience 12 (2): 229–234. February 2009. doi:10.1038/nn.2247. PMID 19079251.
- ↑ "Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri". Nature Neuroscience 11 (6): 631–633. June 2008. doi:10.1038/nn.2120. PMID 18432196.
- ↑ 29.0 29.1 29.2 "Neocortical excitation/inhibition balance in information processing and social dysfunction". Nature 477 (7363): 171–178. July 2011. doi:10.1038/nature10360. PMID 21796121. Bibcode: 2011Natur.477..171Y.
- ↑ "Neural substrates of awakening probed with optogenetic control of hypocretin neurons". Nature 450 (7168): 420–424. November 2007. doi:10.1038/nature06310. PMID 17943086. Bibcode: 2007Natur.450..420A.
- ↑ "Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning". Science 324 (5930): 1080–1084. May 2009. doi:10.1126/science.1168878. PMID 19389999. Bibcode: 2009Sci...324.1080T.
- ↑ "Optical deconstruction of parkinsonian neural circuitry". Science 324 (5925): 354–359. April 2009. doi:10.1126/science.1167093. PMID 19299587. Bibcode: 2009Sci...324..354G.
- ↑ "Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry". Nature 466 (7306): 622–626. July 2010. doi:10.1038/nature09159. PMID 20613723. Bibcode: 2010Natur.466..622K.
- ↑ "Global and local fMRI signals driven by neurons defined optogenetically by type and wiring". Nature 465 (7299): 788–792. June 2010. doi:10.1038/nature09108. PMID 20473285. Bibcode: 2010Natur.465..788L.
- ↑ 35.0 35.1 35.2 "Optical induction of synaptic plasticity using a light-sensitive channel". Nature Methods 4 (2): 139–141. February 2007. doi:10.1038/nmeth988. PMID 17195846.
- ↑ 36.0 36.1 "Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections". Nature Neuroscience 10 (5): 663–668. May 2007. doi:10.1038/nn1891. PMID 17435752.
- ↑ 37.0 37.1 "The columnar and laminar organization of inhibitory connections to neocortical excitatory cells". Nature Neuroscience 14 (1): 100–107. January 2011. doi:10.1038/nn.2687. PMID 21076426.
- ↑ "High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice". Proceedings of the National Academy of Sciences of the United States of America 104 (19): 8143–8148. May 2007. doi:10.1073/pnas.0700384104. PMID 17483470. Bibcode: 2007PNAS..104.8143W.
- ↑ "In-depth activation of channelrhodopsin 2-sensitized excitable cells with high spatial resolution using two-photon excitation with a near-infrared laser microbeam". Biophysical Journal 95 (8): 3916–3926. October 2008. doi:10.1529/biophysj.108.130187. PMID 18621808. Bibcode: 2008BpJ....95.3916M.
- ↑ "Two-photon excitation of channelrhodopsin-2 at saturation". Proceedings of the National Academy of Sciences of the United States of America 106 (35): 15025–15030. September 2009. doi:10.1073/pnas.0907084106. PMID 19706471. Bibcode: 2009PNAS..10615025R.
- ↑ "Two-photon single-cell optogenetic control of neuronal activity by sculpted light". Proceedings of the National Academy of Sciences of the United States of America 107 (26): 11981–11986. June 2010. doi:10.1073/pnas.1006620107. PMID 20543137. Bibcode: 2010PNAS..10711981A.
- ↑ "The Brain Prize 2013: the optogenetics revolution". Trends in Neurosciences 36 (10): 557–560. October 2013. doi:10.1016/j.tins.2013.08.005. PMID 24054067.
- ↑ "Temporal control of immediate early gene induction by light". PLOS ONE 4 (12): e8185. December 2009. doi:10.1371/journal.pone.0008185. PMID 19997631. Bibcode: 2009PLoSO...4.8185S.
- ↑ "High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels". Proceedings of the National Academy of Sciences of the United States of America 108 (18): 7595–7600. May 2011. doi:10.1073/pnas.1017210108. PMID 21504945. Bibcode: 2011PNAS..108.7595B.
- ↑ "Channelrhodopsin-2-XXL, a powerful optogenetic tool for low-light applications". Proceedings of the National Academy of Sciences of the United States of America 111 (38): 13972–13977. September 2014. doi:10.1073/pnas.1408269111. PMID 25201989. Bibcode: 2014PNAS..11113972D.
- ↑ "ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation". Nature Neuroscience 16 (10): 1499–1508. October 2013. doi:10.1038/nn.3502. PMID 23995068.
- ↑ "Independent optical excitation of distinct neural populations". Nature Methods 11 (3): 338–346. March 2014. doi:10.1038/nmeth.2836. PMID 24509633.
- ↑ "Dual-channel circuit mapping reveals sensorimotor convergence in the primary motor cortex". The Journal of Neuroscience 35 (10): 4418–4426. March 2015. doi:10.1523/JNEUROSCI.3741-14.2015. PMID 25762684.
- ↑ "Spike-timing-dependent plasticity rewards synchrony rather than causality". Cerebral Cortex 33 (1): 23–34. December 2022. doi:10.1093/cercor/bhac050. PMID 35203089.
- ↑ "All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins". Nature Methods 11 (8): 825–833. August 2014. doi:10.1038/nmeth.3000. PMID 24952910.
- ↑ "Sensitive red protein calcium indicators for imaging neural activity". eLife 5. March 2016. doi:10.7554/eLife.12727. PMID 27011354.
- ↑ "All-Optical Assay to Study Biological Neural Networks" (in English). Frontiers in Neuroscience 12: 451. 2018. doi:10.3389/fnins.2018.00451. PMID 30026684.
- ↑ "Ultra light-sensitive and fast neuronal activation with the Ca²+-permeable channelrhodopsin CatCh". Nature Neuroscience 14 (4): 513–518. April 2011. doi:10.1038/nn.2776. PMID 21399632.
- ↑ "Calcium-permeable channelrhodopsins for the photocontrol of calcium signalling". Nature Communications 13 (1): 7844. December 2022. doi:10.1038/s41467-022-35373-4. PMID 36543773. Bibcode: 2022NatCo..13.7844F.
- ↑ "Kalium rhodopsins: Natural light-gated potassium channels". bioRxiv: 2021.09.17.460684. 2021-09-17. doi:10.1101/2021.09.17.460684.
- ↑ "WiChR, a highly potassium-selective channelrhodopsin for low-light one- and two-photon inhibition of excitable cells". Science Advances 8 (49): eadd7729. December 2022. doi:10.1126/sciadv.add7729. PMID 36383037. Bibcode: 2022SciA....8D7729V.
- ↑ 57.0 57.1 "Conversion of channelrhodopsin into a light-gated chloride channel". Science 344 (6182): 409–412. April 2014. doi:10.1126/science.1249375. PMID 24674867. Bibcode: 2014Sci...344..409W.
- ↑ 58.0 58.1 "An improved chloride-conducting channelrhodopsin for light-induced inhibition of neuronal activity in vivo". Scientific Reports 5: 14807. October 2015. doi:10.1038/srep14807. PMID 26443033. Bibcode: 2015NatSR...514807W.
- ↑ 59.0 59.1 "Structural foundations of optogenetics: Determinants of channelrhodopsin ion selectivity". Proceedings of the National Academy of Sciences of the United States of America 113 (4): 822–829. January 2016. doi:10.1073/pnas.1523341113. PMID 26699459. Bibcode: 2016PNAS..113..822B.
- ↑ "Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons". Current Biology 18 (15): 1133–1137. August 2008. doi:10.1016/j.cub.2008.06.077. PMID 18682213.
- ↑ "Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice". Nature 451 (7174): 61–64. January 2008. doi:10.1038/nature06445. PMID 18094685. Bibcode: 2008Natur.451...61H.
- ↑ "Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution". PLOS ONE 2 (3): e299. March 2007. doi:10.1371/journal.pone.0000299. PMID 17375185. Bibcode: 2007PLoSO...2..299H.
- ↑ "Multimodal fast optical interrogation of neural circuitry". Nature 446 (7136): 633–639. April 2007. doi:10.1038/nature05744. PMID 17410168. Bibcode: 2007Natur.446..633Z.
- ↑ "NEUROSCIENCE. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics". Science 349 (6248): 647–650. August 2015. doi:10.1126/science.aaa7484. PMID 26113638. Bibcode: 2015Sci...349..647G.
- ↑ "Independent optical excitation of distinct neural populations". Nature Methods 11 (3): 338–346. March 2014. doi:10.1038/nmeth.2836. PMID 24509633.
- ↑ "BiPOLES is an optogenetic tool developed for bidirectional dual-color control of neurons". Nature Communications 12 (1): 4527. July 2021. doi:10.1038/s41467-021-24759-5. PMID 34312384. Bibcode: 2021NatCo..12.4527V.
- ↑ "Optical induction of plasticity at single synapses reveals input-specific accumulation of alphaCaMKII". Proceedings of the National Academy of Sciences of the United States of America 105 (33): 12039–12044. August 2008. doi:10.1073/pnas.0802940105. PMID 18697934. Bibcode: 2008PNAS..10512039Z.
- ↑ "Spike-timing-dependent plasticity rewards synchrony rather than causality". Cerebral Cortex 33 (1): 23–34. February 2022. doi:10.1093/cercor/bhac050. PMID 35203089.
- ↑ "Spike-based indirect training of a spiking neural network-controlled virtual insect". 52nd IEEE Conference on Decision and Control. IEEE Decision and Control. Dec 2013. pp. 6798–6805. doi:10.1109/CDC.2013.6760966. ISBN 978-1-4673-5717-3.
- ↑ "The subcellular organization of neocortical excitatory connections". Nature 457 (7233): 1142–1145. February 2009. doi:10.1038/nature07709. PMID 19151697. Bibcode: 2009Natur.457.1142P.
- ↑ "Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration". Neuron 50 (1): 23–33. April 2006. doi:10.1016/j.neuron.2006.02.026. PMID 16600853.
- ↑ "Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration". Neuron 50 (1): 23–33. April 2006. doi:10.1016/j.neuron.2006.02.026. PMID 16600853.
- ↑ "Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration". Nature Neuroscience 11 (6): 667–675. June 2008. doi:10.1038/nn.2117. PMID 18432197.
- ↑ "Partial recovery of visual function in a blind patient after optogenetic therapy". Nature Medicine 27 (7): 1223–1229. July 2021. doi:10.1038/s41591-021-01351-4. PMID 34031601.
- ↑ "Algae proteins partially restore man's sight". BBC News. 24 May 2021. https://www.bbc.com/news/health-57226572.
- ↑ "Optogenetic stimulation of the auditory pathway". The Journal of Clinical Investigation 124 (3): 1114–1129. March 2014. doi:10.1172/JCI69050. PMID 24509078.
- ↑ "High frequency neural spiking and auditory signaling by ultrafast red-shifted optogenetics". Nature Communications 9 (1): 1750. May 2018. doi:10.1038/s41467-018-04146-3. PMID 29717130. Bibcode: 2018NatCo...9.1750M.
- ↑ "Ultrafast optogenetic stimulation of the auditory pathway by targeting-optimized Chronos". The EMBO Journal 37 (24): e99649. December 2018. doi:10.15252/embj.201899649. PMID 30396994.
Further reading
- "Algal sensory photoreceptors". Annual Review of Plant Biology 59: 167–189. 2008. doi:10.1146/annurev.arplant.59.032607.092847. PMID 18444900. (Naturel function of channelrhodopsins and other photoreceptors in green)
- "In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2". Neuron 54 (2): 205–218. April 2007. doi:10.1016/j.neuron.2007.03.005. PMID 17442243. (Using channelrhodopsin in transgenic mice to study brain circuitry)
- "Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration". Neuron 50 (1): 23–33. April 2006. doi:10.1016/j.neuron.2006.02.026. PMID 16600853. (Using channelrhodopsin potentially to treat blindness)
External links
 |
