Biology:Diaminopimelate decarboxylase
 A cartoon of Methanococcus jannaschii diaminopimelate decarboxylase | |||||||||
| Identifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| EC number | 4.1.1.20 | ||||||||
| CAS number | 9024-75-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
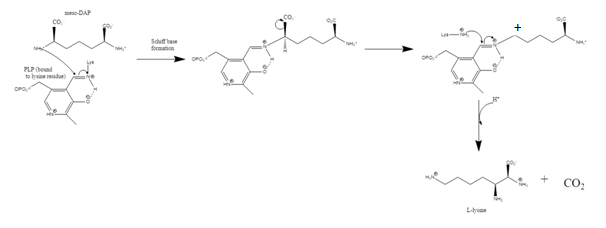
The enzyme diaminopimelate decarboxylase (EC 4.1.1.20) catalyzes the cleavage of carbon-carbon bonds in meso 2,6 diaminoheptanedioate to produce CO2 and L-lysine, the essential amino acid. It employs the cofactor pyridoxal phosphate, also known as PLP, which participates in numerous enzymatic transamination, decarboxylation and deamination reactions.[1]
This enzyme belongs to the family of lyases, specifically the carboxy-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is meso-2,6-diaminoheptanedioate carboxy-lyase (L-lysine-forming).DAP-decarboxylase catalyzes the final step in the meso-diaminopimelate/lysine biosynthetic pathway.[2] Lysine is used for protein synthesis and used in the peptidoglycan layer of Gram-positive bacteria cell walls.[2] This enzyme is not found in humans, but the ortholog is ornithine decarboxylase.[3]
Structure
DAPDC is a PLP-dependent enzyme belonging to the alanine racemase family.[4] This enzyme is generally dimeric with each monomer containing two domains.[5] The first domain is the N-terminal α/β-barrel that binds the PLP to the active site lysine residue.[3][4][5] The second domain is the C-terminal β-sandwich.[4][5] The active site is formed from residues present in both domains resulting in two active sites within the dimer.[5]
DAPDC is stereochemically specific due to the opposing chiralities at each terminus of diaminopimelate.[5] In order for the L-lysine to be generated over D-lysine, decarboxylation must occur at the D-terminus. Whether DAPDC recognizes the terminus or not is dependent on the formation of a Schiff base with PLP.[5]
While the majority of DAPDC found in various species of bacteria have the same basic components, not all species follow the same structure.[3] Some species of bacteria, such as Mycobacterium tuberculosis have been observed as a tetramer.[6] The tetramer is shaped like a ring with the active sites accessible from the inside of the enzyme.[6]
Mechanism
The first step in the mechanism is the same as for all type III PLP-dependent enzymes; the formation of a Schiff base with the substrate amino group.[5] The lysine residue binding PLP to the structure is replaced by diaminopimelate.[4][7] DAPDC then uses the interaction of 3 residues (Arginine, Aspartate, and Glutamate) within the active site to identify the D-stereocenter.[3][7] The DAP is decarboxylated and then stabilized by PLP.[4] It is not clear which general acid protonates after decarboxylation, but there is speculation that the lysine residue is the donor.[7]
Regulation
DAPDC is regulated by the product L-lysine at relatively high concentrations.[3][8] Compounds that are similar to DAP in chemical complexity do not inhibit the reaction, possibly due to the residue rulers creating specific bond angles.[3] Diamines have a stronger inhibitory effect compared to dicarboxylic acids, most likely from interactions with PLP.[3]
Function
Given that there are three pathways to convert aspartate to lysine, this is clearly an essential process for the cell, particularly in building cell walls in Gram-positive bacteria.[2][9] There is no process for producing lysine in humans, but ornithine decarboxylase shares many similarities with DAPDC.[4] Both enzymes use PLP as a cofactor and have similar structures forming the active sites.[7] However, DAPDC differs in that it decarboxylates at the D-stereocenter and is highly stereospecific.[7] These unique features make DAPDC a good candidate for antibacterial studies because potential inhibitors of such an integral step in cell viability would be unlikely to interact with necessary processes within humans.
References
- ↑ "Pyridoxal phosphate". Pubchem. https://pubchem.ncbi.nlm.nih.gov/compound/pyridoxal_phosphate#section=Top.
- ↑ 2.0 2.1 2.2 "Lysine biosynthesis in bacteria: a metallodesuccinylase as a potential antimicrobial target". Journal of Biological Inorganic Chemistry 18 (2): 155–63. February 2013. doi:10.1007/s00775-012-0965-1. PMID 23223968.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 "Dimerization of Bacterial Diaminopimelate Decarboxylase Is Essential for Catalysis". The Journal of Biological Chemistry 291 (18): 9785–95. April 2016. doi:10.1074/jbc.M115.696591. PMID 26921318.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 "Functional classification of amino acid decarboxylases from the alanine racemase structural family by phylogenetic studies". Molecular Biology and Evolution 24 (1): 79–89. January 2007. doi:10.1093/molbev/msl133. PMID 16997906.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 "Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor". Structure 10 (11): 1499–508. November 2002. doi:10.1016/S0969-2126(02)00880-8. PMID 12429091.
- ↑ 6.0 6.1 "The three-dimensional structure of diaminopimelate decarboxylase from Mycobacterium tuberculosis reveals a tetrameric enzyme organisation". Journal of Structural and Functional Genomics 10 (3): 209–17. September 2009. doi:10.1007/s10969-009-9065-z. PMID 19543810.
- ↑ 7.0 7.1 7.2 7.3 7.4 "Analysis of catalytic determinants of diaminopimelate and ornithine decarboxylases using alternate substrates". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1814 (9): 1113–9. September 2011. doi:10.1016/j.bbapap.2011.05.014. PMID 21640851.
- ↑ "Control of lysine biosynthesis in Bacillus subtilis: inhibition of diaminopimelate decarboxylase by lysine". Journal of Bacteriology 121 (1): 20–8. January 1975. doi:10.1128/JB.121.1.20-28.1975. PMID 234936.
- ↑ Dogovski, Con; Atkinson, Sarah C.; Dommaraju, Sudhir R.; Dobson, Renwick C. J.; Perugini, Matthew A. (2009). "Lysine biosynthesis in bacteria – an unchartered pathway for novel antibiotic design". Biotechnology XI: 146–166. http://www.eolss.net/sample-chapters/c17/E6-58-10-09.pdf.
Further reading
- "Diaminopimelic acid decarboxylase in pyridoxin-deficient Escherichia coli". Biochimica et Biophysica Acta 16 (3): 442–3. March 1955. doi:10.1016/0006-3002(55)90257-2. PMID 14378182.
 |