Biology:Enterobacter virus CC31
| Enterobacter virus CC31 | |
|---|---|

| |
| Basic 2D and 3D structures of Enterobacter virus CC31 | |
| Virus classification | |
| Group: | Group I (dsDNA)
|
| Order: | |
| Family: | |
| Subfamily: | |
| Genus: | Cc31virus
|
| Species: | Enterobacter virus CC31
|
Enterobacter virus CC31 is a dsDNA bacteriophage of the subfamily Tevenvirinae responsible for infecting the bacteria family of Enterobactericeae.[1] It is one of two discovered viruses of the genus Cc31virus, diverging away from the previously discovered T4virus, as a clonal complex (CC).[2][3][4] CC31 was first isolated from Escherichia coli B strain S/6/4 and is primarily associated with Escherichia, even though is named after Enterobacter.[5][6]
Viral Classification and Structure
Enterobacter virus CC31 is a dsDNA virus lacking an RNA intermediate. The dsDNA is contained within an icosahedral capsid of proteins, but the virus lacks an envelope. It is in the order Caudovirales, being that it is a bacteriophage with a protein sheath and tail used to infect host cells. Caudovirales genetic material is contained within an icosahedral capsid resting on top of the sheath and tail.[7] CC31 is in the Myoviridae family due to its lack of envelope, linear genome, and long, helical, permanent tail subunits. Tevenvirinae is the relatively large (43 genera) subfamily CC13 falls underneath. Finally, Cc13virus is the relatively new genus associated with Enterobacter virus CC31.[8]
Genome
Enterobacter virus CC31 has a double stranded linear DNA (dsDNA) genome consisting of 165,540 nucleotide base pairs. The bases account for 287 genes that are capable of making 279 different proteins using 8 tRNAs.[6][9] 93% of the genetic material is homologous with Enterobacter virus PG7, the other Tevenvirinae, and 74% of the material is homologous with the close relative Enterobacteria phage T-4.[10] 120 new open reading frames (ORFs) were identified across the base pairs that were added to the Enterobacteria phage pan-genome. It is currently the only phage, that is not a T-even bacteriophage, capable of encoding for glycosyltransferases.[6]
Genetic Modification
The CC31 is capable of integrating its genetic material with the host's. This stage, known as the lysogenic cycle, halts particle formation, and allows for the virus's genetic material to amplify across many subsequent bacterium generations.[11] The integration of the viral DNA happens non-homologously, forming bubbles of single stranded DNA. As DNA replication, crossing over, and cellular division occurs, the viral DNA is reshuffled with cellular DNA. This results in horizontal gene transfer between the virus and cell, resulting in the evolution of the virus and the bacterium. The viral DNA also develops minute deletions, immunity regions, and silent genetic regions due to the non-homologous binding of the DNA. These change in virus' genetic material can either inhibit or promote future replication.[12]
Pathogenesis
In order for viral genomic replication to occur, CC31 needs to enter the host, Escherichia coli. Due to the lack of an outer envelope, the virion must find an alternate way to enter its host. It does this by penetrating the membrane of the bacterium with its tail region. First, the long tail fibers ejecting from the top of the base plate attach to the cell membrane of the bacterium. The small tail fibers underneath base plate then attach to the membrane, initiating a conformational change of the base plate to a six-point star from the previous hexagon conformation. This forces the sheath to undergo a conformational change to contract and effectively stretching the membrane. While this is occurring, the rigid tube underneath the sheath remains stagnant to push against the membrane. Digestion of the inner membrane occurs with the assistance of the tail's lysozymes. As this continues, the inner tube breaks through the membrane and allows the viral DNA to flow into the cytoplasm of the bacterium.[13]
Lytic Cycle
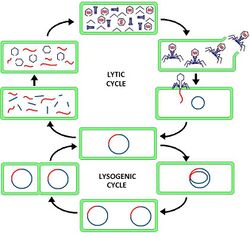
Once inside the cell, replication can begin. The virus starts by breaking down all E. coli genetic material. This is known as the lytic cycle. The virus can now occupy the cell of the E. coli without being inhibited by proteins or enzymes of the host. The CC30 genetic material is then capable of using the residue E. coli proteins to assist with viral replication. Enterobacter virus CC31 has most of the genes responsible for coding proteins to induce gene expression and replication: endonuclease, RNA polymerase, DNA polymerase, RNA primase, DNA ligase, topoisomerase, and DNA helicase.[14] Therefore, CC31 does not require access to E. coli's nucleus and does not have to wait for mitosis to occur in order to act in a parasitic fashion. This allows for rapid formation of virus particles within the empty cell. DNA is amplified, while proteins are made for the virion construction. Protein subunits combine into domains to make the individual components of the virion. The viral particles begin to combine once overcrowding of the cell occurs. The E. coli cell will lyse due to over-population, allowing the virions to burst out of the cell and move on to the next host.[12][15][16]
Lysogenic Cycle
The virus is also capable of taking a different pathway to replicate its DNA known as the lysogenic cycle. Instead of destroying the cellular DNA, the viral DNA integrates itself within it with the assistance of integrase, to become a silent provirus. This integration forms non-homologous single stranded bubbles of viral DNA. These regions are susceptible to damage, resulting in frame shifts, minute deletions, immunity regions, and silent genetic regions. These modifications, patterned with horizontal gene transfer between E. coli DNA and CC30 DNA, allow for evolution to occur for the virus and bacterium. This patience illustrated by the virus allows for it to amplify its genetic material significantly through many E. coli generations. Also, as the integrated viral DNA is translated into mRNA, proteins are synthesized and are readily available for future virion formation. Once a stressor is induced onto the cell, the integration weakens and subsequently releases the viral genetic material. The virus now enters the lytic cycle and begins replication in the numerous bacteria cells it now occupies.[15][16][17] As the lytic cycle progresses and the virions begin to infect new cells, the acquired E. coli genetic material can now be transduced into other cells as the bacteriophage reenters the lysogenic cycle.[12]
Interactions with Enterobacteriaceae
Beta-lactamase (blaCMY-2) is an enzyme responsible for providing antibiotic resistance to penicillins, cephalosporins, and carbapenems. Hydrolysis of the antibiotics by blaCMY-2 results in the resistance.[18] This enzyme is present and expressed in Salmonella choleraesuis, a bacterium primarily associated with infecting cattle and poultry. This gene presents a global issue for the consumption of food products. The antibiotic resistance of Salmonella makes it difficult to treat these infections if they were to inflict humans. The genetic material coding for the blaCMY-2 enzyme was not ancestrally part of the bacterium's genome, but was acquired by the IncI1 plasmid.[19]
E. coli local to the human's GI tract have acquired this same antibiotic resistance using the blaCMY-2 enzyme. The sequence coding for the blaCMY-2 gene is a derivative of the IncI1 plasmid. The distinct divergence of E. coli and S. choleraesuis removes the possibility of this occurrence being from plasma transmission between the species.[20] The acquisition of these sequences is a result of Enterobacter virus CC31's ability to influence gene transduction.
With the plasma integrated into the bacterium DNA and the CC31 in the lysogenic cycle, genetic material is exchanged across subsequent generations. Random crossing over and gene transfer results in heterozygosity of the bacteria and prophage. After a stressor is induced onto the cells and the virus enters the lytic cycle to eventually lyse the cell, CC31 is free to roam around the body to infect a different bacteria species and undergo random gene transfer once again. As this continues, variable gene fragments from the plasma and virus are transferred. These random gene transfers have resulted in adoption of the IncI1 and a new antibiotic resistant E. coli.[4]
References
- ↑ Krupovic, Mart; Dutilh, Bas E.; Adriaenssens, Evelien M.; Wittmann, Johannes; Vogensen, Finn K.; Sullivan, Mathew B.; Rumnieks, Janis; Prangishvili, David et al. (2016-04-01). "Taxonomy of prokaryotic viruses: update from the ICTV bacterial and archaeal viruses subcommittee" (in en). Archives of Virology 161 (4): 1095–1099. doi:10.1007/s00705-015-2728-0. ISSN 0304-8608. https://link.springer.com/article/10.1007/s00705-015-2728-0.
- ↑ taxonomy. "Taxonomy Browser". https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=1913650&lvl=3&keep=1&srchmode=1&unlock.
- ↑ taxonomy. "Taxonomy Browser". https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=1198136&lvl=3&lin=f&keep=1&srchmode=1&unlock.
- ↑ 4.0 4.1 Bielak, E.M., Hasman, H. and Aarestrup, F.M., 2012. Diversity and epidemiology of plasmids from Enterobacteriaceae from human and non-human reservoirs (Doctoral dissertation, Technical University of DenmarkDanmarks Tekniske Universitet, National Food InstituteFødevareinstituttet, Division of Epidemiology and Microbial GenomicsAfdeling for Epidemiologi og Genomisk Mikrobiologi).
- ↑ "Enterobacter phage CC31". http://www.genome.jp/virushostdb/709484.
- ↑ 6.0 6.1 6.2 Petrov, Vasiliy M.; Ratnayaka, Swarnamala; Nolan, James M.; Miller, Eric S.; Karam, Jim D. (2010-10-28). "Genomes of the T4-related bacteriophages as windows on microbial genome evolution". Virology Journal 7: 292. doi:10.1186/1743-422X-7-292. ISSN 1743-422X. https://doi.org/10.1186/1743-422X-7-292.
- ↑ Boundless. "Boundless Microbiology | Simple Book Publishing" (in en-US). https://courses.lumenlearning.com/boundless-microbiology/.
- ↑ taxonomy. "Taxonomy Browser". https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1913650&lvl=3&lin=f&keep=1&srchmode=1&unlock.
- ↑ "Enterobacter phage CC31 (ID 4102) - Genome - NCBI". https://www.ncbi.nlm.nih.gov/genome/?term=txid709484%5BOrganism:noexp%5D.
- ↑ "Enterobacteria phage CC31, complete genome - Nucleotide - NCBI". https://www.ncbi.nlm.nih.gov/nucleotide/284177830?report=genbank&log$=nucltop&blast_rank=1&RID=ZFCTGWHZ015.
- ↑ khanacademymedicine (2015-01-20), Viral replication: lytic vs lysogenic | Cells | MCAT | Khan Academy, https://www.youtube.com/watch?v=J4BN4dARpio, retrieved 2017-11-02
- ↑ 12.0 12.1 12.2 Jane,, Flint, S.. Principles of virology. Racaniello, V. R. (Vincent R.),, Rall, Glenn F.,, Skalka, Anna M.,, Enquist, L. W. (Lynn W.), (4th ed.). Washington, DC. ISBN 9781555819347. OCLC 914445879. https://www.worldcat.org/oclc/914445879.
- ↑ Taylor, Nicholas M. I.; Prokhorov, Nikolai S.; Guerrero-Ferreira, Ricardo C.; Shneider, Mikhail M.; Browning, Christopher; Goldie, Kenneth N.; Stahlberg, Henning; Leiman, Petr G.. "Structure of the T4 baseplate and its function in triggering sheath contraction". Nature 533 (7603): 346–352. doi:10.1038/nature17971. http://www.nature.com/doifinder/10.1038/nature17971.
- ↑ "txid1913656[Organism - Protein - NCBI"]. https://www.ncbi.nlm.nih.gov/protein?term=txid1913656%5BOrganism%5D.
- ↑ 15.0 15.1 (in en) Viral replication: lytic vs lysogenic, https://www.khanacademy.org/test-prep/mcat/cells/viruses/v/viral-replicaiton-lytic-vs-lysogenic, retrieved 2017-11-02
- ↑ 16.0 16.1 Jane,, Flint, S.. Principles of virology. Racaniello, V. R. (Vincent R.),, Rall, Glenn F.,, Skalka, Anna M.,, Enquist, L. W. (Lynn W.), (4th ed.). Washington, DC. ISBN 9781555819330. OCLC 914445879. https://www.worldcat.org/oclc/914445879.
- ↑ (in en) Retroviruses, https://www.khanacademy.org/test-prep/mcat/cells/viruses/v/retroviruses, retrieved 2017-11-02
- ↑ Brouwer, Michael S. M.; Bossers, Alex; Harders, Frank; Essen-Zandbergen, Alieda van; Mevius, Dik J.; Smith, Hilde E. (2014-08-28). "Complete Genome Sequences of IncI1 Plasmids Carrying Extended-Spectrum β-Lactamase Genes" (in en). Genome Announcements 2 (4): e00859–14. doi:10.1128/genomea.00859-14. ISSN 2169-8287. PMID 25169863. http://genomea.asm.org/content/2/4/e00859-14.
- ↑ Call, Douglas R.; Singer, Randall S.; Meng, Da; Broschat, Shira L.; Orfe, Lisa H.; Anderson, Janet M.; Herndon, David R.; Kappmeyer, Lowell S. et al. (2010-02-01). "blaCMY-2-Positive IncA/C Plasmids from Escherichia coli and Salmonella enterica Are a Distinct Component of a Larger Lineage of Plasmids" (in en). Antimicrobial Agents and Chemotherapy 54 (2): 590–596. doi:10.1128/aac.00055-09. ISSN 0066-4804. PMID 19949054. PMC 2812137. http://aac.asm.org/content/54/2/590.
- ↑ Tagg, Kaitlin A.; Iredell, Jonathan R.; Partridge, Sally R. (2014-08-01). "Complete Sequencing of IncI1 Sequence Type 2 Plasmid pJIE512b Indicates Mobilization of blaCMY-2 from an IncA/C Plasmid" (in en). Antimicrobial Agents and Chemotherapy 58 (8): 4949–4952. doi:10.1128/aac.02773-14. ISSN 0066-4804. PMID 24890591. PMC 4135994. http://aac.asm.org/content/58/8/4949.



