Biology:Pan-genome

In the fields of molecular biology and genetics, a pan-genome (pangenome or supragenome) is the entire set of genes from all strains within a clade. More generally, it is the union of all the genomes of a clade.[2][3][4][5] The pan-genome can be broken down into a "core pangenome" that contains genes present in all individuals, a "shell pangenome" that contains genes present in two or more strains, and a "cloud pangenome" that contains genes only found in a single strain.[3][4][6] Some authors also refer to the cloud genome as "accessory genome" containing 'dispensable' genes present in a subset of the strains and strain-specific genes.[2][3][4] Note that the use of the term 'dispensable' has been questioned, at least in plant genomes, as accessory genes play "an important role in genome evolution and in the complex interplay between the genome and the environment".[5] The field of study of pangenomes is called pangenomics.[2]
The genetic repertoire of a bacterial species is much larger than the gene content of an individual strain. [7] Some species have open (or extensive) pangenomes, while others have closed pangenomes.[2] For species with a closed pan-genome, very few genes are added per sequenced genome (after sequencing many strains), and the size of the full pangenome can be theoretically predicted. Species with an open pangenome have enough genes added per additional sequenced genome that predicting the size of the full pangenome is impossible.[4] Population size and niche versatility have been suggested as the most influential factors in determining pan-genome size.[2]
Pangenomes were originally constructed for species of bacteria and archaea, but more recently eukaryotic pan-genomes have been developed, particularly for plant species. Plant studies have shown that pan-genome dynamics are linked to transposable elements.[8][9][10][11] The significance of the pan-genome arises in an evolutionary context, especially with relevance to metagenomics,[12] but is also used in a broader genomics context.[13] An open access book reviewing the pangenome concept and its implications, edited by Tettelin and Medini, was published in the spring of 2020.[14]
Etymology
The term 'pangenome' was defined with its current meaning by Tettelin et al. in 2005;[2] it derives 'pan' from the Greek word παν, meaning 'whole' or 'everything', while the genome is a commonly used term to describe an organism's complete genetic material. Tettelin et al. applied the term specifically to bacteria, whose pangenome "includes a core genome containing genes present in all strains and a dispensable genome composed of genes absent from one or more strains and genes that are unique to each strain."[2]
Parts of the pangenome
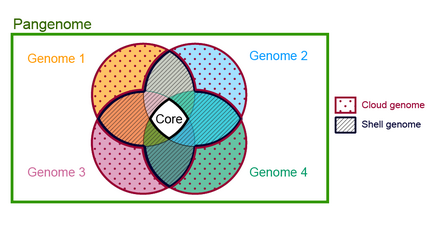
Core
Is the part of the pangenome that is shared by every genome in the tested set. Some authors have divided the core pangenome in hard core, those families of homologous genes that has at least one copy of the family shared by every genome (100% of genomes) and the soft core or extended core,[15] those families distributed above a certain threshold (90%). In a study that involves the pangenomes of Bacillus cereus and Staphylococcus aureus, some of them isolated from the international space station, the thresholds used for segmenting the pangenomes were as follows: "Cloud," "Shell," and "Core" corresponding to gene families with presence in <10%, 10 to 95%, and >95% of the genomes, respectively.[16]
The core genome size and proportion to the pangenome depends on several factors, but it is especially dependent on the phylogenetic similarity of the considered genomes. For example, the core of two identical genomes would also be the complete pangenome. The core of a genus will always be smaller than the core genome of a species. Genes that belong to the core genome are often related to house keeping functions and primary metabolism of the lineage, nevertheless, the core gene can also contain some genes that differentiate the species from other species of the genus, i.e. that may be related pathogenicity to niche adaptation.[17]
Shell
Is the part of the pangenome shared by the majority of the genomes in a pangenome.[18] There is not a universally accepted threshold to define the shell genome, some authors consider a gene family as part of the shell pangenome if it shared by more than 50% of the genomes in the pangenome.[19] A family can be part of the shell by several evolutive dynamics, for example by gene loss in a lineage where it was previously part of the core genome, such is the case of enzymes in the tryptophan operon in Actinomyces,[20] or by gene gain and fixation of a gene family that was previously part of the dispensable genome such is the case of trpF gene in several Corynebacterium species.[21]
Cloud
The cloud genome consists of those gene families shared by a minimal subset of the genomes in the pangenome,[22] it includes singletons or genes present in only one of the genomes. It is also known as the peripheral genome, or accessory genome. Gene families in this category are often related to ecological adaptation.[citation needed]
Classification

The pan-genome can be somewhat arbitrarily classified as open or closed based on the alpha value of the Heap law: [23][15]
- Number of gene families.
- Number of genomes.
- Constant of proportionality.
- Exponent calculated in order to adjust the curve of number of gene families vs new genome.
if then the pangenome is considered open. if then the pangenome is considered closed.
Usually, the pangenome software can calculate the parameters of the Heap law that best describe the behavior of the data.
Open pangenome
An open pangenome occurs when in one taxonomic lineage keeps increasing the number of new gene families and this increment does not seem to be asymptotic regardless how many new genomes are added to the pangenome. Escherichia coli is an example of a species with an open pangenome. Any E. coli genome size is in the range of 4000-5000 genes and the pangenome size estimated for this species with approximately 2000 genomes is composed by 89,000 different gene families.[24] The pangenome of the domain bacteria is also considered to be open.
Closed Pangenome
A closed pangenome occurs in a lineage when only few gene families are added when new genomes are incorporated into the pangenome analysis, and the total amount of gene families in the pangenome seem to be asymptotic to one number. It is believed that parasitism and species that are specialists in some ecological niche tend to have closed pangenomes. Staphylococcus lugdunensis is an example of a commensal bacteria with closed pan-genome.[25]
History
Pangenome
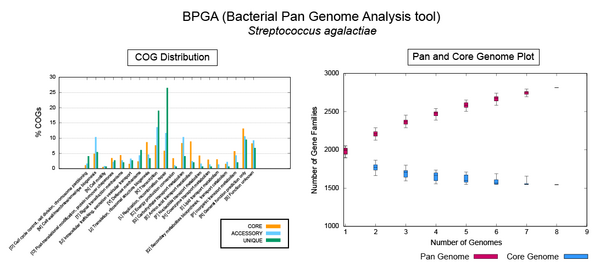
The original pangenome concept was developed by Tettelin et al.[2] when they analyzed the genomes of eight isolates of Streptococcus agalactiae, where they described a core genome shared by all isolates, accounting for approximately 80% of any single genome, plus a dispensable genome consisting of partially shared and strain-specific genes. Extrapolation suggested that the gene reservoir in the S. agalactiae pan-genome is vast and that new unique genes would continue to be identified even after sequencing hundreds of genomes.[2] The pangenome comprises the entirety of the genes discovered in the sequenced genomes of a given microbial species and it can change when new genomes are sequenced and incorporated into the analysis.[citation needed]

The pangenome of a genomic lineage accounts for the intra lineage gene content variability. Pangenome evolves due to: gene duplication, gene gain and loss dynamics and interaction of the genome with mobile elements that are shaped by selection and drift.[26] Some studies point that prokaryotes pangenomes are the result of adaptive, not neutral evolution that confer species the ability to migrate to new niches.[27]
Supergenome
The supergenome can be thought of as the real pangenome size if all genomes from a species were sequenced.[28] It is defined as all genes accessible for being gained by a certain species. It cannot be calculated directly but its size can be estimated by the pangenome size calculated from the available genome data. Estimating the size of the cloud genome can be troubling because of its dependence on the occurrence of rare genes and genomes. In 2011 genomic fluidity was proposed as a measure to categorize the gene-level similarity among groups of sequenced isolates. [29] In some lineages the supergenomes did appear infinite,[30] as is the case of the Bacteria domain.[31]
Metapangenome
'Metapangenome' has been defined as the outcome of the analysis of pangenomes in conjunction with the environment where the abundance and prevalence of gene clusters and genomes are recovered through shotgun metagenomes.[32] The combination of metagenomes with pangenomes, also referred to as "metapangenomics", reveals the population-level results of habitat-specific filtering of the pangenomic gene pool.[33]
Other authors consider that Metapangenomics expands the concept of pangenome by incorporating gene sequences obtained from uncultivated microorganisms by a metagenomics approach. A metapangenome comprises both sequences from metagenome-assembled genomes (MAGs) and from genomes obtained from cultivated microorganisms.[34] Metapangenomics has been applied to assess diversity of a community, microbial niche adaptation, microbial evolution, functional activities, and interaction networks of the community.[35] The Anvi'o platform developed a workflow that integrates analysis and visualization of metapangenomes by generating pangenomes and study them in conjunction with metagenomes.[32]
Examples
Prokaryote pangenome
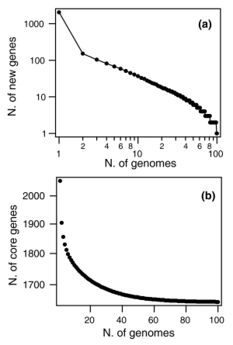
In 2018, 87% of the available whole genome sequences were bacteria fueling researchers interest in calculating prokaryote pangenomes at different taxonomic levels.[22] In 2015, the pangenome of 44 strains of Streptococcus pneumoniae bacteria shows few new genes discovered with each new genome sequenced (see figure). In fact, the predicted number of new genes dropped to zero when the number of genomes exceeds 50 (note, however, that this is not a pattern found in all species). This would mean that S. pneumoniae has a 'closed pangenome'.[37] The main source of new genes in S. pneumoniae was Streptococcus mitis from which genes were transferred horizontally. The pan-genome size of S. pneumoniae increased logarithmically with the number of strains and linearly with the number of polymorphic sites of the sampled genomes, suggesting that acquired genes accumulate proportionately to the age of clones.[36] Another example of prokaryote pan-genome is Prochlorococcus, the core genome set is much smaller than the pangenome, which is used by different ecotypes of Prochlorococcus.[38] Open pan-genome has been observed in environmental isolates such as Alcaligenes sp.[39] and Serratia sp.,[40] showing a sympatric lifestyle. Nevertheless, open pangenome is not exclusive to free living microorganisms, a 2015 study on Prevotella bacteria isolated from humans, compared the gene repertoires of its species derived from different body sites of human. It also reported an open pan-genome showing vast diversity of gene pool.[41]
Archaea also have some pangenome studies. Halobacteria pangenome shows the following gene families in the pangenome subsets: core (300), variable components (Softcore: 998, Cloud:36531, Shell:11784).[42]
Eukaryote pangenome
Eukaryote organisms such as fungi, animals and plants have also shown evidence of pangenomes. In four fungi species whose pangenome has been studied, between 80 and 90% of gene models were found as core genes. The remaining accessory genes were mainly involved in pathogenesis and antimicrobial resistance.[43]
In animals, the human pangenome is being studied. In 2010 a study estimated that a complete human pan-genome would contain ~19–40 Megabases of novel sequence not present in the extant reference human genome.[44] The Human Pangenome consortium has the goal to acknowledge the human genome diversity. In 2023, a draft human pangenome reference was published.[45] It is based on 47 genomes from persons of varied ethnicity.[45] Plans are underway for an improved reference capturing still more biodiversity from a still wider sample.[45]
Among plants, there are examples of pangenome studies in model species, both diploid [9] and polyploid,[10] and a growing list of crops.[46][47] Pangenomes have shown promise as a tool in plant breeding by accounting for structural variants and SNPs in non-reference genomes, which helps to solve the problem of missing heritability that persists in genome wide association studies.[48] An emerging plant-based concept is that of pan-NLRome, which is the repertoire of nucleotide-binding leucine-rich repeat (NLR) proteins, intracellular immune receptors that recognize pathogen proteins and confer disease resistance.[49]
Virus pangenome
Virus does not necessarily have genes extensively shared by clades such as is the case of 16S in bacteria, and therefore the core genome of the full Virus Domain is empty. Nevertheless, several studies have calculated the pangenome of some viral lineages. The core genome from six species of pandoraviruses comprises 352 gene families only 4.7% of the pangenome, resulting in an open pangenome.[50]
Data structures
The number of sequenced genomes is continuously growing "simply scaling up established bioinformatics pipelines will not be sufficient for leveraging the full potential of such rich genomic data sets".[51] Pangenome graphs are emerging data structures designed to represent pangenomes and to efficiently map reads to them. They have been reviewed by Eizenga et al. [52]
Software tools

As interest in pangenomes increased, there have been several software tools developed to help analyze this kind of data. To start a pangenomic analysis the first step is the homogenization of genome annotation.[23] The same software should be used to annotate all genomes used, such as GeneMark[53] or RAST.[54] In 2015, a group reviewed the different kinds of analyses and tools a researcher may have available.[55] There are seven kinds of software developed to analyze pangenomes: Those dedicated to cluster homologous genes; identify SNPs; plot pangenomic profiles; build phylogenetic relationships of orthologous genes/families of strains/isolates; function-based searching; annotation and/or curation; and visualization.[55]
The two most cited software tools for pangenomic analysis at the end of 2014[55] were Panseq[56] and the pan-genomes analysis pipeline (PGAP).[57] Other options include BPGA – A Pan-Genome Analysis Pipeline for prokaryotic genomes,[58] GET_HOMOLOGUES,[59] Roary.[60] and PanDelos.[61] In 2015 a review focused on prokaryote pangenomes[62] and another for plant pan-genomes were published.[63] Among the first software packages designed for plant pangenomes were PanTools.[64] and GET_HOMOLOGUES-EST.[11][59] In 2018 panX was released, an interactive web tool that allows inspection of gene families evolutionary history.[65] panX can display an alignment of genomes, a phylogenetic tree, mapping of mutations and inference about gain and loss of the family on the core-genome phylogeny. In 2019 OrthoVenn 2.0 [66] allowed comparative visualization of families of homologous genes in Venn diagrams up to 12 genomes. In 2023, BRIDGEcerealwas developed to survey and graph indel-based haplotypes from pan-genome through a gene model ID.[67]

In 2020 Anvi'o[1] was available as a multiomics platform that contains pangenomic and metapangenomic analyses as well as visualization workflows. In Anvi'o, genomes are displayed in concentrical circles and each radius represents a gene family, allowing for comparison of more than 100 genomes in its interactive visualization. In 2020, a computational comparison of tools for extracting gene-based pangenomic contents (such as GET_HOMOLOGUES, PanDelos, Roary, and others) has been released.[68] Tools were compared from a methodological perspective, analyzing the causes that lead a given methodology to outperform other tools. The analysis was performed by taking into account different bacterial populations, which are synthetically generated by changing evolutionary parameters. Results show a differentiation of the performance of each tool that depends on the composition of the input genomes. Again in 2020, several tools introduced a graphical representation of the pangenomes showing the contiguity of genes (PPanGGOLiN,[46] Panaroo[65]).
See also
- Metagenomics
- Pathogenomics
- Quasispecies
References
- ↑ 1.0 1.1 "Community-led, integrated, reproducible multi-omics with anvi'o". Nature Microbiology 6 (1): 3–6. January 2021. doi:10.1038/s41564-020-00834-3. PMID 33349678.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 Tettelin, Hervé; Masignani, Vega; Cieslewicz, Michael J.; Donati, Claudio; Medini, Duccio; Ward, Naomi L.; Angiuoli, Samuel V.; Crabtree, Jonathan et al. (2005-09-27). "Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial "pan-genome"" (in en). Proceedings of the National Academy of Sciences 102 (39): 13950–13955. doi:10.1073/pnas.0506758102. ISSN 0027-8424. PMID 16172379. Bibcode: 2005PNAS..10213950T.
- ↑ 3.0 3.1 3.2 "The microbial pan-genome". Current Opinion in Genetics & Development 15 (6): 589–94. December 2005. doi:10.1016/j.gde.2005.09.006. PMID 16185861.
- ↑ 4.0 4.1 4.2 4.3 "Ten years of pan-genome analyses". Current Opinion in Microbiology 23: 148–54. February 2015. doi:10.1016/j.mib.2014.11.016. PMID 25483351.
- ↑ 5.0 5.1 "Structural variation and genome complexity: is dispensable really dispensable?". Current Opinion in Plant Biology 18: 31–36. April 2014. doi:10.1016/j.pbi.2014.01.003. PMID 24548794.
- ↑ "Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer". Biology Direct 7: 46. December 2012. doi:10.1186/1745-6150-7-46. PMID 23241446.
- ↑ "The bacterial pan-genome:a new paradigm in microbiology". Int Microbiol 13 (2): 45–57. 2010. doi:10.2436/20.1501.01.110. PMID 20890839. https://pubmed.ncbi.nlm.nih.gov/20890839/.
- ↑ "Transposable elements and the plant pan-genomes". Current Opinion in Plant Biology 10 (2): 149–55. April 2007. doi:10.1016/j.pbi.2007.02.001. PMID 17300983.
- ↑ 9.0 9.1 "Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure". Nature Communications 8 (1): 2184. December 2017. doi:10.1038/s41467-017-02292-8. PMID 29259172. Bibcode: 2017NatCo...8.2184G.
- ↑ 10.0 10.1 "Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors". Nature Communications 11 (1): 3670. July 2020. doi:10.1038/s41467-020-17302-5. PMID 32728126. Bibcode: 2020NatCo..11.3670G.
- ↑ 11.0 11.1 "Analysis of Plant Pan-Genomes and Transcriptomes with GET_HOMOLOGUES-EST, a Clustering Solution for Sequences of the Same Species". Frontiers in Plant Science 8: 184. February 2017. doi:10.3389/fpls.2017.00184. PMID 28261241.
- ↑ "Biogeography of the Sulfolobus islandicus pan-genome". Proceedings of the National Academy of Sciences of the United States of America 106 (21): 8605–10. May 2009. doi:10.1073/pnas.0808945106. PMID 19435847. Bibcode: 2009PNAS..106.8605R.
- ↑ "De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae". Genome Research 19 (2): 294–305. February 2009. doi:10.1101/gr.083311.108. PMID 19015323.
- ↑ (in en-gb) The Pangenome. 2020. doi:10.1007/978-3-030-38281-0. ISBN 978-3-030-38280-3. http://library.oapen.org/bitstream/20.500.12657/37707/1/2020_Book_ThePangenome.pdf.
- ↑ 15.0 15.1 "Calculating orthologs in bacteria and Archaea: a divide and conquer approach". PLOS ONE 6 (12): e28388. 2011. doi:10.1371/journal.pone.0028388. PMID 22174796. Bibcode: 2011PLoSO...628388H.
- ↑ "Pangenomic Approach To Understanding Microbial Adaptations within a Model Built Environment, the International Space Station, Relative to Human Hosts and Soil". mSystems 4 (1): e00281-18. 2019. doi:10.1128/mSystems.00281-18. PMID 30637341.
- ↑ "Pangenome-wide and molecular evolution analyses of the Pseudomonas aeruginosa species". BMC Genomics 17 (45): 45. January 2016. doi:10.1186/s12864-016-2364-4. PMID 26754847.
- ↑ "Standard operating procedure for computing pangenome trees". Standards in Genomic Sciences 2 (1): 135–41. January 2010. doi:10.4056/sigs.38923. PMID 21304685.
- ↑ "EvoMining reveals the origin and fate of natural product biosynthetic enzymes". Microbial Genomics 5 (12): e000260. December 2019. doi:10.1099/mgen.0.000260. PMID 30946645.
- ↑ "Evolution of substrate specificity in a retained enzyme driven by gene loss". eLife 6 (6): e22679. March 2017. doi:10.7554/eLife.22679. PMID 28362260.
- ↑ "Evolution of substrate specificity in a recipient's enzyme following horizontal gene transfer". Molecular Biology and Evolution 30 (9): 2024–34. September 2013. doi:10.1093/molbev/mst115. PMID 23800623.
- ↑ 22.0 22.1 "A Review of Pangenome Tools and Recent Studies". The Pangenome. 2020. pp. 89–112. doi:10.1007/978-3-030-38281-0_4. ISBN 978-3-030-38280-3.
- ↑ 23.0 23.1 "First Steps in the Analysis of Prokaryotic Pan-Genomes". Bioinformatics and Biology Insights 14: 1177932220938064. 2020. doi:10.1177/1177932220938064. PMID 32843837.
- ↑ "Insights from 20 years of bacterial genome sequencing". Functional & Integrative Genomics 15 (2): 141–61. March 2015. doi:10.1007/s10142-015-0433-4. PMID 25722247.
- ↑ "Comparative genomic analysis of Staphylococcus lugdunensis shows a closed pan-genome and multiple barriers to horizontal gene transfer". BMC Genomics 19 (1): 621. August 2018. doi:10.1186/s12864-018-4978-1. PMID 30126366.
- ↑ "The Ecology and Evolution of Pangenomes". Current Biology 29 (20): R1094–R1103. October 2019. doi:10.1016/j.cub.2019.08.012. PMID 31639358.
- ↑ "Why prokaryotes have pangenomes". Nature Microbiology 2 (4): 17040. March 2017. doi:10.1038/nmicrobiol.2017.40. PMID 28350002. http://eprints.whiterose.ac.uk/113972/39/McInerney_McNally_O%27Connell_NatMicro.pdf.
- ↑ "The Turbulent Network Dynamics of Microbial Evolution and the Statistical Tree of Life". Journal of Molecular Evolution 80 (5–6): 244–50. June 2015. doi:10.1007/s00239-015-9679-7. PMID 25894542. Bibcode: 2015JMolE..80..244K.
- ↑ "Genomic fluidity: an integrative view of gene diversity within microbial populations". BMC Genomics 12 (12): 32. January 2011. doi:10.1186/1471-2164-12-32. PMID 21232151.
- ↑ "Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes". BMC Biology 12 (66): 66. August 2014. doi:10.1186/s12915-014-0066-4. PMID 25141959.
- ↑ "Estimating the size of the bacterial pan-genome". Trends in Genetics 25 (3): 107–10. March 2009. doi:10.1016/j.tig.2008.12.004. PMID 19168257.
- ↑ 32.0 32.1 Delmont, TO; Eren, AM (2018). "Linking pangenomes and metagenomes: the Prochlorococcus metapangenome.". PeerJ 6: e4320. doi:10.7717/peerj.4320. PMID 29423345.
- ↑ Utter, Daniel R.; Borisy, Gary G.; Eren, A. Murat; Cavanaugh, Colleen M.; Mark Welch, Jessica L. (2020-12-16). "Metapangenomics of the oral microbiome provides insights into habitat adaptation and cultivar diversity". Genome Biology 21 (1): 293. doi:10.1186/s13059-020-02200-2. PMID 33323129.
- ↑ "Meta-Pangenome: At the Crossroad of Pangenomics and Metagenomics". The Pangenome: Diversity, Dynamics and Evolution of Genomes. Springer. 2020. pp. 205–218. doi:10.1007/978-3-030-38281-0_9. ISBN 978-3-030-38281-0. https://www.ncbi.nlm.nih.gov/books/NBK558817/.
- ↑ "Integrating pan-genome with metagenome for microbial community profiling". Computational and Structural Biotechnology Journal 19: 1458–1466. 2021. doi:10.1016/j.csbj.2021.02.021. PMID 33841754.
- ↑ 36.0 36.1 "Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species". Genome Biology 11 (10): R107. 2010. doi:10.1186/gb-2010-11-10-r107. PMID 21034474.
- ↑ "The bacterial pangenome as a new tool for analysing pathogenic bacteria". New Microbes and New Infections 7: 72–85. September 2015. doi:10.1016/j.nmni.2015.06.005. PMID 26442149.
- ↑ "Patterns and implications of gene gain and loss in the evolution of Prochlorococcus". PLOS Genetics 3 (12): e231. December 2007. doi:10.1371/journal.pgen.0030231. PMID 18159947.
- ↑ "Genome sequencing and analysis of Alcaligenes faecalis subsp. phenolicus MB207". Scientific Reports 8 (1): 3616. 2018. doi:10.1038/s41598-018-21919-4. PMID 29483539. Bibcode: 2018NatSR...8.3616B.
- ↑ Basharat Z, Yasmin A (2016). "Pan-genome Analysis of the Genus Serratia". arXiv:1610.04160 [q-bio.GN].
- ↑ "Divergences in gene repertoire among the reference Prevotella genomes derived from distinct body sites of human". BMC Genomics 16 (153): 153. March 2015. doi:10.1186/s12864-015-1350-6. PMID 25887946.
- ↑ "Pan-genome analysis and ancestral state reconstruction of class halobacteria: probability of a new super-order". Scientific Reports 10 (1): 21205. December 2020. doi:10.1038/s41598-020-77723-6. PMID 33273480. Bibcode: 2020NatSR..1021205G.
- ↑ "Pan-genome analyses of model fungal species". Microbial Genomics 5 (2). February 2019. doi:10.1099/mgen.0.000243. PMID 30714895.
- ↑ "Building the sequence map of the human pan-genome". Nat Biotechnol 28 (1): 57–63. 2010. doi:10.1038/nbt.1596. PMID 19997067. https://www.nature.com/articles/nbt.1596.
- ↑ 45.0 45.1 45.2 Liao, Wen-WeiExpression error: Unrecognized word "et". (May 2023). "A draft human pangenome reference". Nature 617 (7960): 312–324. doi:10.1038/s41586-023-05896-x. PMID 37165242.
- ↑ 46.0 46.1 "The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor". Nature Genetics 51 (6): 1044–51. May 2019. doi:10.1038/s41588-019-0410-2. PMID 31086351.
- ↑ "The barley pan-genome reveals the hidden legacy of mutation breeding". Nature 588 (7837): 284–9. Nov 2020. doi:10.1038/s41586-020-2947-8. PMID 33239781. Bibcode: 2020Natur.588..284J.
- ↑ Zhou, Yao; Zhang, Zhiyang; Bao, Zhigui; Li, Hongbo; Lyu, Yaqing; Zan, Yanjun; Wu, Yaoyao; Cheng, Lin et al. (8 July 2022). "Graph pangenome captures missing heritability and empowers tomato breeding" (in en). Nature 606 (7914): 527–534. doi:10.1038/s41586-022-04808-9. ISSN 1476-4687. PMID 35676474.
- ↑ "A Species-Wide Inventory of NLR Genes and Alleles in Arabidopsis thaliana". Cell 178 (5): 1260–72. August 2019. doi:10.1016/j.cell.2019.07.038. PMID 31442410.
- ↑ "A Large Open Pangenome and a Small Core Genome for Giant Pandoraviruses". Frontiers in Microbiology 9 (9): 1486. 2018. doi:10.3389/fmicb.2018.01486. PMID 30042742.
- ↑ "Computational pan-genomics: status, promises and challenges". Briefings in Bioinformatics 19 (1): 118–135. January 2018. doi:10.1093/bib/bbw089. PMID 27769991.
- ↑ "Pangenome Graphs". Annual Review of Genomics and Human Genetics 21: 139–162. August 2020. doi:10.1146/annurev-genom-120219-080406. PMID 32453966.
- ↑ "GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions". Nucleic Acids Research 29 (12): 2607–18. June 2001. doi:10.1093/nar/29.12.2607. PMID 11410670.
- ↑ "The RAST Server: rapid annotations using subsystems technology". BMC Genomics 9 (9): 75. February 2008. doi:10.1186/1471-2164-9-75. PMID 18261238.
- ↑ 55.0 55.1 55.2 "A brief review of software tools for pangenomics". Genomics, Proteomics & Bioinformatics 13 (1): 73–6. February 2015. doi:10.1016/j.gpb.2015.01.007. PMID 25721608.
- ↑ "Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions". BMC Bioinformatics 11 (1): 461. September 2010. doi:10.1186/1471-2105-11-461. PMID 20843356.
- ↑ "PGAP: pan-genomes analysis pipeline". Bioinformatics 28 (3): 416–8. February 2012. doi:10.1093/bioinformatics/btr655. PMID 22130594.
- ↑ "BPGA- an ultra-fast pan-genome analysis pipeline". Scientific Reports 6 (24373): 24373. April 2016. doi:10.1038/srep24373. PMID 27071527. Bibcode: 2016NatSR...624373C.
- ↑ 59.0 59.1 "GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis". Applied and Environmental Microbiology 79 (24): 7696–701. December 2013. doi:10.1128/AEM.02411-13. PMID 24096415. Bibcode: 2013ApEnM..79.7696C.
- ↑ "Roary: rapid large-scale prokaryote pan genome analysis". Bioinformatics 31 (22): 3691–3. November 2015. doi:10.1093/bioinformatics/btv421. PMID 26198102.
- ↑ "PanDelos: a dictionary-based method for pan-genome content discovery". BMC Bioinformatics 19 (Suppl 15): 437. November 2018. doi:10.1186/s12859-018-2417-6. PMID 30497358.
- ↑ "Inside the Pan-genome - Methods and Software Overview". Current Genomics 16 (4): 245–52. August 2015. doi:10.2174/1389202916666150423002311. PMID 27006628.
- ↑ "Towards plant pangenomics". Plant Biotechnology Journal 14 (4): 1099–105. April 2016. doi:10.1111/pbi.12499. PMID 26593040. http://espace.library.uq.edu.au/view/UQ:383261/UQ383261_OA.pdf.
- ↑ "PanTools: Representation, Storage and Exploration of Pan-Genomic Data". Bioinformatics 32 (17): i487–i493. September 2016. doi:10.1093/bioinformatics/btw455. PMID 27587666.
- ↑ 65.0 65.1 "panX: pan-genome analysis and exploration". Nucleic Acids Research 46 (1): e5. January 2018. doi:10.1093/nar/gkx977. PMID 29077859.
- ↑ "OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species". Nucleic Acids Research 47 (W1): W52–W58. July 2019. doi:10.1093/nar/gkz333. PMID 31053848.
- ↑ Zhang, Bosen; Huang, Haiyan; Tibbs-Cortes, Laura E.; Vanous, Adam; Zhang, Zhiwu; Sanguinet, Karen; Garland-Campbell, Kimberly A.; Yu, Jianming et al. (2023). "Streamline unsupervised machine learning to survey and graph indel-based haplotypes from pan-genomes" (in en). Molecular Plant 16 (6): 975–978. doi:10.1016/j.molp.2023.05.005. PMID 37202927.
- ↑ "Challenges in gene-oriented approaches for pangenome content discovery". Briefings in Bioinformatics 22 (3). 2020. doi:10.1093/bib/bbaa198. ISSN 1477-4054. PMID 32893299.
 |
