Biology:FANCM
 Generic protein structure example |
| Fanconi anemia, complementation group M | |
|---|---|
| Identifiers | |
| Symbol | FANCM |
| Alt. symbols | KIAA1596 |
| NCBI gene | 57697 |
| HGNC | 23168 |
| OMIM | 609644 |
| PDB | 4BXO |
| RefSeq | XM_048128 |
| UniProt | Q8IYD8 |
| Other data | |
| EC number | 3.6.1.- |
| Locus | Chr. 14 q21.3 |
Fanconi anemia, complementation group M, also known as FANCM is a human gene.[1][2] It is an emerging target in cancer therapy, in particular cancers with specific genetic deficiencies.[3][4]
Function
The protein encoded by this gene, FANCM displays DNA binding against fork structures[5] and an ATPase activity associated with DNA branch migration. It is believed that FANCM in conjunction with other Fanconi anemia- proteins repair DNA at stalled replication forks, and stalled transcription structures called R-loops.[6][7]
The structure of the C-terminus of FANCM (amino acids 1799-2048), bound to a partner protein FAAP24, reveals how the protein complex recognises branched DNA.[5] A structure of amino acids 675-790 of FANCM reveal how the protein binds duplex DNA through a remodeling of the MHF1:MHF2 histone-like protein complex.

Disease association
Bi-allelic mutations in the FANCM gene were originally associated with Fanconi anemia, although several individuals with FANCM deficiency do not appear to have the disorder.[9][10][11] Mono-allelic FANCM mutations are associated with breast cancer risk and especially with risk of developing ER-negative and TNBC disease subtypes.[12][13][14] A founder mutation in the Scandinavian population is also associated with a higher than average frequency of triple negative breast cancer in heterozygous carriers.[15] FANCM carriers also have elevated levels of Ovarian cancer and other solid tumours[16]
FANCM as a therapeutic target in ALT cancer
Expression and activity of FANCM, is essential for the viability of cancers using Alternative Lengthening of Telomeres (ALT-associated cancers).[17][18][19] Several other synthetic lethal interactions have been observed for FANCM that may widen the targetability of the protein in therapeutic use.[17][4]
There are several potential ways in which FANCM activity could be targeted as an anti-cancer agent. In the context of ALT, one of the best targets may be a peptide domain of FANCM called MM2. Ectopic MM2 peptide (that acts as a dominant decoy) was sufficient to inhibit colony formation of ALT-associated cancer cells, but not telomerase-positive cancer cells.[18] This peptide works as a dominant interfering binder to RMI1:RMI2, and sequesters another DNA repair complex called the Bloom Syndrome complex away from FANCM.[7] As with FANCM depletion, this induces death through a “hyper-ALT” phenotype. An in vitro high-throughput screen for small molecule inhibitors of MM2-RMI1:2 interaction lead to the discovery of PIP-199.[20] This experimental drug also showed some discriminatory activity in killing of ALT-cells, compared to telomerase-positive cells.[18]
Meiosis
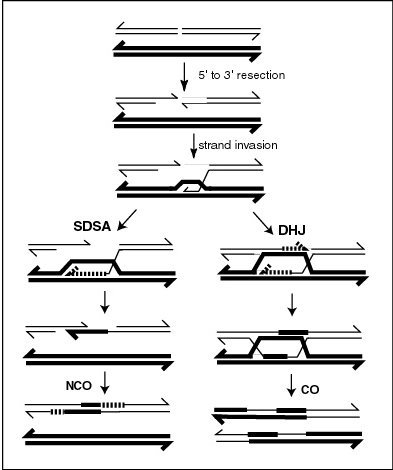
Recombination during meiosis is often initiated by a DNA double-strand break (DSB). During recombination, sections of DNA at the 5' ends of the break are cut away in a process called resection. In the strand invasion step that follows, an overhanging 3' end of the broken DNA molecule then "invades" the DNA of a homologous chromosome that is not broken forming a displacement loop (D-loop). After strand invasion, the further sequence of events may follow either of two main pathways leading to a crossover (CO) or a non-crossover (NCO) recombinant (see Genetic recombination and Homologous recombination). The pathway leading to a NCO is referred to as synthesis dependent strand annealing (SDSA).
In the plant Arabidopsis thaliana FANCM helicase antagonizes the formation of CO recombinants during meiosis, thus favoring NCO recombinants.[21] The FANCM helicase is required for genome stability in humans and yeast, and is a major factor limiting meiotic CO formation in A. thaliana.[22] A pathway involving another helicase, RECQ4A/B, also acts independently of FANCM to reduce CO recombination.[21] These two pathways likely act by unwinding different joint molecule substrates (e.g. nascent versus extended D-loops; see Figure).
Only about 4% of DSBs in A. thaliana are repaired by CO recombination;[22] the remaining 96% are likely repaired mainly by NCO recombination. Sequela-Arnaud et al.[21] suggested that CO numbers are restricted because of the long-term costs of CO recombination, that is, the breaking up of favorable genetic combinations of alleles built up by past natural selection.
In the fission yeast Schizosaccharomyces pombe, FANCM helicase also directs NCO recombination during meiosis.[23]
References
- ↑ "Prediction of the coding sequences of unidentified human genes. XVIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Research 7 (4): 273–81. August 2000. doi:10.1093/dnares/7.4.271. PMID 10997877.
- ↑ "A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M". Nature Genetics 37 (9): 958–63. September 2005. doi:10.1038/ng1626. PMID 16116422.
- ↑ "Breaking the end: Target the replication stress response at the ALT telomeres for cancer therapy". Molecular & Cellular Oncology 4 (6): e1360978. 2017-11-02. doi:10.1080/23723556.2017.1360978. PMID 29209649.
- ↑ 4.0 4.1 "ALT control, delete: FANCM as an anti-cancer target in Alternative Lengthening of Telomeres". Nucleus 10 (1): 221–230. October 2019. doi:10.1080/19491034.2019.1685246. PMID 31663812.
- ↑ 5.0 5.1 5.2 "Architecture and DNA recognition elements of the Fanconi anemia FANCM-FAAP24 complex". Structure 21 (9): 1648–58. September 2013. doi:10.1016/j.str.2013.07.006. PMID 23932590.
- ↑ "The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks". Molecular Cell 29 (1): 141–8. January 2008. doi:10.1016/j.molcel.2007.11.032. PMID 18206976.
- ↑ 7.0 7.1 "FANCM connects the genome instability disorders Bloom's Syndrome and Fanconi Anemia". Molecular Cell 36 (6): 943–53. December 2009. doi:10.1016/j.molcel.2009.12.006. PMID 20064461.
- ↑ "The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder". Annual Review of Biophysics 43: 257–78. 2014. doi:10.1146/annurev-biophys-051013-022737. PMID 24773018.
- ↑ "A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome". Molecular and Cellular Biology 23 (10): 3417–26. May 2003. doi:10.1128/MCB.23.10.3417-3426.2003. PMID 12724401.
- ↑ "Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia". Genetics in Medicine 20 (4): 458–463. April 2018. doi:10.1038/gim.2017.124. PMID 28837157.
- ↑ "Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility". Genetics in Medicine 20 (4): 452–457. April 2018. doi:10.1038/gim.2017.123. PMID 28837162. https://ddd.uab.cat/pub/artpub/2018/182643/genmed_a2018v20p452.pdf.
- ↑ "FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor". Human Molecular Genetics 24 (18): 5345–55. September 2015. doi:10.1093/hmg/ddv251. PMID 26130695.
- ↑ "Association Between Loss-of-Function Mutations Within the FANCM Gene and Early-Onset Familial Breast Cancer". JAMA Oncology 3 (9): 1245–48. September 2017. doi:10.1001/jamaoncol.2016.5592. PMID 28033443.
- ↑ "The FANCM:p.Arg658* truncating variant is associated with risk of triple-negative breast cancer". npj Breast Cancer 5 (38): 38. November 2019. doi:10.1038/s41523-019-0127-5. PMID 31700994.
- ↑ "Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer". Proceedings of the National Academy of Sciences of the United States of America 111 (42): 15172–7. October 2014. doi:10.1073/pnas.1407909111. PMID 25288723. Bibcode: 2014PNAS..11115172K.
- ↑ "FANCM as a likely high grade serous ovarian cancer susceptibility gene". Oncotarget 8 (31): 50930–50940. August 2017. doi:10.18632/oncotarget.15871. PMID 28881617.
- ↑ 17.0 17.1 "FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres". Proceedings of the National Academy of Sciences of the United States of America 114 (29): E5940–E5949. July 2017. doi:10.1073/pnas.1708065114. PMID 28673972. Bibcode: 2017PNAS..114E5940P.
- ↑ 18.0 18.1 18.2 "The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT)". Nature Communications 10 (1): 2252. May 2019. doi:10.1038/s41467-019-10180-6. PMID 31138797. Bibcode: 2019NatCo..10.2252L.
- ↑ "FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops". Nature Communications 10 (1): 2253. May 2019. doi:10.1038/s41467-019-10179-z. PMID 31138795. Bibcode: 2019NatCo..10.2253S.
- ↑ "A High-Throughput Screening Strategy to Identify Protein-Protein Interaction Inhibitors That Block the Fanconi Anemia DNA Repair Pathway". Journal of Biomolecular Screening 21 (6): 626–33. July 2016. doi:10.1177/1087057116635503. PMID 26962873.
- ↑ 21.0 21.1 21.2 "Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM". Proceedings of the National Academy of Sciences of the United States of America 112 (15): 4713–8. April 2015. doi:10.1073/pnas.1423107112. PMID 25825745. Bibcode: 2015PNAS..112.4713S.
- ↑ 22.0 22.1 "FANCM limits meiotic crossovers". Science 336 (6088): 1588–90. June 2012. doi:10.1126/science.1220381. PMID 22723424. Bibcode: 2012Sci...336.1588C.
- ↑ "The fission yeast FANCM ortholog directs non-crossover recombination during meiosis". Science 336 (6088): 1585–8. June 2012. doi:10.1126/science.1220111. PMID 22723423. Bibcode: 2012Sci...336.1585L.
External links
- FANCM protein, human at the US National Library of Medicine Medical Subject Headings (MeSH)
 |
