Biology:KIAA2012
 Generic protein structure example |
KIAA2012 is a protein which, in humans, is encoded by the KIAA2012 gene. KIAA2012 is expressed at very low levels throughout the body, but it is primarily expressed in the ovary, lungs, and brain.[1]
Gene

KIAA2012 is located on the positive sense strand at position 2q33.1.[2] KIAA2012 has 24 exons, and it spans 131,934 bases including introns. No aliases or common names are used in addition to KIAA2012.
Gene level regulation
Within the promoter region of KIAA2012, there is a highly conserved transcription factor binding site that has no common SNPs.[3] The RFX transcription factors, more specifically RFX1-6, bind to this highly conserved region and regulates cellular specialization and differentiation.[4] The image below shows the promoter region of KIAA2012 with the highly conserved RFX1-6 binding site.[3]

mRNA
KIAA2012 is expressed differentially in the body at low levels. Of this overall low expression, KIAA2012 is expressed most highly in the brain, lungs, and ovary.[1][5] KIAA2012 is expressed at lower levels in the liver, trachea, and testes.[6][7][8]
Protein
Unmodified KIAA2012 is 1,181 amino acids in length, has a molecular weight of 136 kdal, and an isoelectric pH around 8.[9][10]
Internal features
KIAA2012 is rich in glutamic acid and glutamine, and it is poor in valine.[9] There is also one mixed charge cluster between amino acids 951–1118.[11] There is one Domain of Unknown Function (DUF 4670) within KIAA2012 spanning from amino acid 635 to amino acid 1137.[2] Different than the whole KIAA2012, DUF 4670 is also rich in arginine and poor in glycine and phenylalanine.[9]

Structure
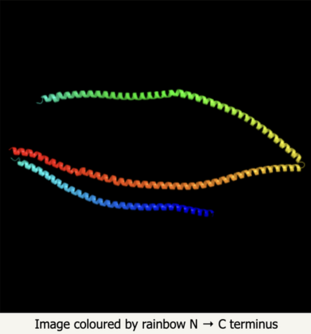
The secondary structure of KIAA2012 consists primarily of alpha helices. On the left, a high confidence prediction of the secondary structure is shown. On the right, the entire 3-D structure is shown, showing how the alpha helices fold to form the entire KIAA2012 protein.

Post-translational modification
KIAA2012 has a highly conserved cGMP-dependent protein kinase binding domain. These cGMP-dependent protein kinases (PRKG) are a part of the NO/cGMP signaling pathway, and they are important factors in many signal transduction processes.[12] Additionally, there are many potential sites for phosphorylation, SUMOylation, and myristoylation. In instances where KIAA2012 is post-translationally modified in these ways, the resulting charge, structure, function, and sub-cellular localization can be altered.[13][14]
Sub-cellular Localization
Proteins tagged with localization signals will be transported to various regions of the cell. KIAA2012 contains nuclear localization signal sequences, which are short stretches of amino acids that moderate transportation of nuclear proteins to the nucleus.[15] Shown in the table below, human KIAA2012 and two orthologs are listed with confidence values of where in the cell KIAA2012 is localized.[16]
| Nuclear | Plasma Membrane | Cytoskeletal | Mitochondrial | Cytoplasmic | Secretory Vesicles | |
|---|---|---|---|---|---|---|
| Human | 82% | 4% | 9% | 4% | --- | --- |
| Sardinian Tree Frog | 78% | 9% | 9% | 4% | --- | --- |
| Zebrafish | 74% | 9% | 4% | --- | 9% | 4% |
Function
KIAA2012 has predicted protein interactions with STAG2 and SMC1A.[17] STAG2 encodes a subunit of cohesion complexes used to regulate sister chromatid separation during cell division.[18] SMC1A is an important part of functional kinetochores due to its role in the multiprotein cohesion complex required for sister chromatid cohesion.[19] Because KIAA2012 is localized in the nucleus and interacts with STAG2 and SMC1A, it's role as a protein surrounds DNA manipulation or cell division.
| Protein Name | Aliases | Location |
|---|---|---|
| SMC1A | SMC1, SMCB, CDLS2, SB1.8, SMC1L1, DXS423E, SMC1alpha, RP6-29D12.1 | Xp11.22[19] |
| STAG2 | SA2, SA-2, SCC3B, bA517O1.1, RP11-517O1.1 | Xq25[18] |
Homology and evolution
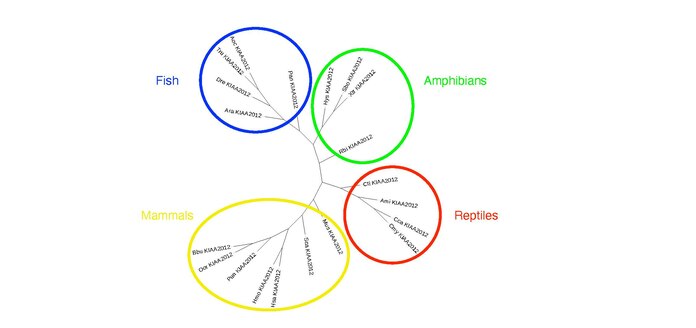
Twenty organisms with a KIAA2012 ortholog are shown below, and they are sorted by date of divergence and sequence identity. There were no orthologs found in birds, but ortholog versions of KIAA2012 exist in mammals, reptiles, amphibians, and fish. An unrooted phylogenetic tree showing each taxonomic group and their divergence patterns can be found below the ortholog table.
| Genus & Species | Common Name | Date of Divergenve (MYA) | Accession # | Sequence Length | % Identity | % Similarity |
| Homo sapiens | Human | 0 | NM_001277372.4 | 1181 | 100 | 100 |
| Hylobates moloch | Silvery Gibbon | 19.5 | XP_032610815 | 1181 | 94.2 | 96.4 |
| Sciurus carolinensis | Gray Squirrel | 87 | XP_047398902 | 1130 | 64.1 | 72.3 |
| Mus caroli | Mouse | 87 | XP_029333762 | 1160 | 61.3 | 72 |
| Panthera uncia | Snow Leopard | 94 | XP_049471125 | 1180 | 75.3 | 82.7 |
| Orcinus orca | Killer Whale | 94 | XP_033285753 | 1172 | 74.5 | 82.7 |
| Bubalus bubalis | Water Buffalo | 94 | XP_006080602 | 1185 | 72.9 | 81.1 |
| Alligator mississippiensis | American Alligator | 319 | XP_059583055 | 1325 | 37.1 | 49.7 |
| Caretta caretta | Loggerhead Turtle | 319 | XP_048725054 | 1329 | 37 | 51.4 |
| Chelonia mydas | Green Sea Turtle | 319 | XP_037768210 | 1325 | 36.8 | 50.9 |
| Crotalus tigris | Tiger Rattlesnake | 319 | XP_039210533 | 1220 | 32.4 | 46.9 |
| Xenopus tropicalis | Western Clawed Frog | 352 | XP_031749269 | 1339 | 31.9 | 45.8 |
| Rhinatrema bivittatum | Two-Lined Caecilian | 352 | XP_029462137 | 1499 | 30.6 | 44.8 |
| Spea bombifrons | Plains Spadefoot Toad | 352 | XP_053326593 | 1436 | 29.6 | 44 |
| Hyla sarda | Sardinian Tree Frog | 352 | XP_056391303 | 1428 | 29.5 | 44.9 |
| Protopterus annectens | West African Lungfish | 408 | XP_043931036 | 1412 | 29.8 | 45 |
| Takifugu rubripes | Japanese Puffer | 429 | XP_029701411 | 1129 | 25.3 | 39.4 |
| Danio rerio | Zebrafish | 429 | XP_009302807 | 1484 | 24.9 | 37.1 |
| Anarrhichthys ocellatus | Wolf Eel | 429 | XP_031729884 | 1204 | 23.9 | 37.3 |
| Amblyraja radiata | Thorny Skate | 462 | XP_032880336 | 1392 | 25.7 | 40.5 |
Clinical significance
There are several genome-wide association studies that report traits associated variations in KIAA2012. The reported traits with the highest number of associations are heel bone mineral density, taste liking measurement, educational attainment, lung function, and height.[20] Additionally, KIAA2012 is down regulated in women with polycystic ovary syndrome (PCOS) compared to women without PCOS.[21]
References
- ↑ 1.0 1.1 Fagerberg, Linn; Hallstrom, Bjorn M; Oksvold, Per; Kampf, Caroline; Djureinovic, Dijana; Odeberg, Jacob; Habuka, Masato; Tahmasebpoor, Simin et al. (5 Dec 2013). "Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics". Mol Cell Proteomics 13 (2): 397–406. doi:10.1074/mcp.M113.035600. PMID 24309898.
- ↑ 2.0 2.1 "KIAA2012 [Homo sapiens (human)"]. National Library of Medicine. https://www.ncbi.nlm.nih.gov/gene/100652824.
- ↑ 3.0 3.1 "USCS Genomics Institute". https://genome.ucsc.edu/cgi-bin/hgc.
- ↑ Sugiaman-Trapman, Debora; Vitezic, Morana; Jouhilahti, Eeva-Mari; Mathelier, Anthony; Lauter, Gilbert; Misra, Sougat; Daub, Carsten O; Kere, Juha et al. (6 March 2018). "Characterization of the human RFX transcription factor family by regulatory and target gene analysis". BMC Genomics 19 (181): 181. doi:10.1186/s12864-018-4564-6. PMID 29510665.
- ↑ "Illumina bodyMap2 Transcriptome". BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJEB2445/.
- ↑ Szabo, Linda; Morey, Robert; Palpant, Nathan J; Wang, Peter L; Afari, Nastaran; Jiang, Chuan; Parast, Mana M; Murry, Charles E et al. (16 June 2015). "Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development". Genome Biol 16 (1): 126. doi:10.1186/s13059-015-0690-5. PMID 26076956.
- ↑ Duff, Michael O; Olson, Sara; Wei, Xintao; Garrett, Sandra C; Osman, Ahmad; Bolisetty, Mohan; Plocik, Alex; Celniker, Susan E et al. (13 May 2015). "Genome-wide identification of zero nucleotide recursive splicing in Drosophila". Nature 521 (7552): 376–379. doi:10.1038/nature14475. PMID 25970244. Bibcode: 2015Natur.521..376D.
- ↑ "Tissue Expression Type -- KIAA2012". https://www.proteinatlas.org/ENSG00000182329-KIAA2012/tissue.
- ↑ 9.0 9.1 9.2 "SAPS Results". European Bioinformatic Institute. https://www.ebi.ac.uk/Tools/seqstats/saps/.
- ↑ Tokmakov, Alexander A.; Kurotani, Atsushi; Sato, Ken-Ichi (2021). "Protein pI and Intracellular Localization". Frontiers in Molecular Biosciences 8. doi:10.3389/fmolb.2021.775736. PMID 34912847.
- ↑ Zhu, Z Y; Karlin, S (1996). "Clusters of Charges Residues in Protein Three-Dimensional Structures". Proceedings of the National Academy of Sciences (Proceedings of the National Academy of Sciences of the United States of America) 93 (16): 8350–8355. doi:10.1073/pnas.93.16.8350. PMID 8710874. Bibcode: 1996PNAS...93.8350Z.
- ↑ Wolfertstetter, Stefanie; Huettner, Johannes P; Schlossmann, Jens (7 Feb 2013). "cGMP-Dependent Kinase Inhibitors in Health and Disease". Pharmaceuticals 6 (2): 269–286. doi:10.3390/ph6020269. PMID 24275951.
- ↑ Maejima, Yasuhiro; Sadoshima, Junichi (24 Sep 2014). "SUMOylation: A Novel Protein Quality Control Modifier in the Heart". Circulation Research 115 (8): 686–689. doi:10.1161/CIRCRESAHA.114.304989. PMID 25258400.
- ↑ Nestler, Eric J; Greengard, Paul (1999). "Protein Phosphorylation is of Fundamental Importance in Biological Regulation". Basic Neurochemistry: Molecular, Cellular and Medical Aspects 6. https://www.ncbi.nlm.nih.gov/books/NBK28063/. Retrieved 10 Dec 2023.
- ↑ Cokol, Murat; Nair, Rajesh; Rost, Burkhard (15 Nov 2000). "Finding nuclear localization signals". EMBO Reports 1 (5): 411–415. doi:10.1093/embo-reports/kvd092. PMID 11258480.
- ↑ "YLoc". https://abi-services.cs.uni-tuebingen.de/yloc/webloc.cgi?id=9298182912ca8acce5493407aff435a5.
- ↑ "KIAA2012 Results Summary". https://thebiogrid.org/1530199.
- ↑ 18.0 18.1 "STAG2 cohesion complex component". National Library of Medicine. https://www.ncbi.nlm.nih.gov/gene/10735.
- ↑ 19.0 19.1 "SMC1A - structural maintenance of chromosome 1A (human)". National Library of Medicine. https://www.ncbi.nlm.nih.gov/gene/8243#gene-expression.
- ↑ "The NHGRI-EBI Catalog of human genome-wide association studies". https://www.ebi.ac.uk/gwas/.
- ↑ Hiam, Danielle; Simar, David; Laker, Rhianna; Altintas, Ali; Gibson-Helm, Melanie; Fletcher, Elly; Moreno-Asso, Alba; Treqin, Adam J et al. (12 Dec 2019). "Epigenetic Reprogramming of Immune Cells in Women With PCOS Impact Genes Controlling Reproductive Function". The Journal of Clinical Endocrinology & Metabolism 104 (12): 6155–6170. doi:10.1210/jc.2019-01015. PMID 31390009.
 |
