Biology:Methyl-accepting chemotaxis proteins
| MCPsignal | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | MCPsignal | ||||||||
| Pfam | PF00015 | ||||||||
| Pfam clan | CL0510 | ||||||||
| InterPro | IPR004089 | ||||||||
| PROSITE | PDOC00465 | ||||||||
| SCOP2 | 1qu7 / SCOPe / SUPFAM | ||||||||
| CDD | cd11386 | ||||||||
| |||||||||
| Methyl-accepting chemotaxis proteins | |||||||||
|---|---|---|---|---|---|---|---|---|---|
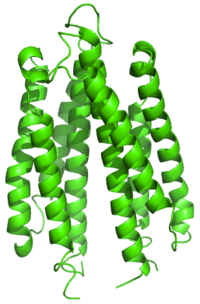 Ribbon diagram of the S. typhimurium aspartate receptor ligand binding domain[2] | |||||||||
| Identifiers | |||||||||
| Symbol | Methyl-accepting chemotaxis proteins (MCP) | ||||||||
| Pfam | PF02203 | ||||||||
| InterPro | IPR004090 | ||||||||
| SMART | TarH | ||||||||
| SCOP2 | 1lih / SCOPe / SUPFAM | ||||||||
| CDD | cd00181 | ||||||||
| |||||||||
The methyl-accepting chemotaxis proteins (MCP, also aspartate receptor) are a family of transmembrane receptors that mediate chemotactic response in certain enteric bacteria, such as Salmonella enterica enterica and Escherichia coli.[3] These methyl-accepting chemotaxis receptors are one of the first components in the sensory excitation and adaptation responses in bacteria, which act to alter swimming behaviour upon detection of specific chemicals. Use of the MCP allows bacteria to detect concentrations of molecules in the extracellular matrix so that the bacteria may smooth swim or tumble accordingly. If the bacterium detects rising levels of attractants (nutrients) or declining levels of repellents (toxins), the bacterium will continue swimming forward, or smooth swimming. If the bacterium detects declining levels of attractants or rising levels of repellents, the bacterium will tumble and re-orient itself in a new direction. In this manner, a bacterium may swim towards nutrients and away from toxins[4]
Evolution
There are many different types of bacterial 60 kDa transmembrane receptors, which share similar topology and signalling mechanisms. They possess three domains: a periplasmic ligand-binding domain, two transmembrane segments, and a cytoplasmic domain. The structure of the ligand-binding domain comprises a closed or partly opened, four-helical bundle with a left-handed twist. The difference in the sequence of the ligand-binding domain between receptors reflects the different ligand specificities. Binding of the ligand causes a conformational change that is transmitted across the membrane to the cytoplasmic activation domain.[5]
Environmental diversity gives rise to diversity in bacterial signalling receptors, and consequently there are many genes encoding MCPs.[6] For example, there are four well-characterised MCPs found in Escherichia coli: Tar (taxis towards aspartate and maltose, away from nickel and cobalt), Tsr (taxis towards serine, away from leucine, indole and weak acids), Trg (taxis towards galactose and ribose) and Tap (taxis towards dipeptides).
Structure
MCPs share similar structure and signalling mechanism. MCPs form dimers. Three dimers of MCP spontaneously form trimers. Trimers are complexed by CheA and CheW into hexagonal lattices. MCPs either bind ligands directly or interact with ligand-binding proteins, transducing the signal to downstream signalling proteins in the cytoplasm. Most MCPs contain: (a) an N-terminal signal peptide that is a transmembrane alpha-helix in the mature protein; (b) a poorly-conserved periplasmic receptor (ligand-binding) domain; (c) a transmembrane alpha-helix; (d) generally one or more HAMP domains and (e) a highly conserved C-terminal cytoplasmic domain that interacts with downstream signalling components. The C-terminal domain contains the methylated glutamate residues.
MCPs undergo two covalent modifications: deamidation and reversible methylation at a number of glutamate residues. Attractants increase the level of methylation, while repellents decrease it. The methyl groups are added by the methyl-transferase CheR and are removed by the methylesterase CheB.
Function
Binding a ligand causes a conformational change in the MCP receptor which translates down the hairpin structure and inhibits its sensor kinase. At the tip of the hairpin are two proteins that associate to the MCP: CheW and CheA. CheA acts as the sensor kinase. CheA has kinase activity and autophosphorylates itself on a histidyl residue when activated by the MCP. CheW is believed to be a transducer of the signal from the MCP to CheA. Activated CheA transfers its phosphoryl group to CheY, a response regulator. Phosphorylated CheY phosphorylates the basal body FliM which is connected to the flagellum. Phosphorylation of the basal body acts as a flagellar switch and changes the direction of rotation of the flagellum. This change in direction allows for alternation between smooth swimming and tumbling which biases the bacterial random walk towards attractant.
References
- ↑ Ferris, H. U.; Zeth, K.; Hulko, M.; Dunin-Horkawicz, S.; Lupas, A. N. (2014). "Axial helix rotation as a mechanism for signal regulation inferred from the crystallographic analysis of the E. Coli serine chemoreceptor". Journal of Structural Biology 186 (3): 349–356. doi:10.1016/j.jsb.2014.03.015. PMID 24680785.
- ↑ PDB: 1VLT; "High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor". J Mol Biol 262 (2): 186–201. 1996. doi:10.1006/jmbi.1996.0507. PMID 8831788.; rendered with PyMOL
- ↑ "High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor". J. Mol. Biol. 262 (2): 186–201. 1996. doi:10.1006/jmbi.1996.0507. PMID 8831788.
- ↑ "Changing the specificity of a bacterial chemoreceptor". J. Mol. Biol. 355 (5): 923–32. February 2006. doi:10.1016/j.jmb.2005.11.025. PMID 16359703.
- ↑ "Propagating conformational changes over long (and short) distances in proteins". Proc. Natl. Acad. Sci. U.S.A. 98 (17): 9517–9520. 2001. doi:10.1073/pnas.161239298. PMID 11504940. PMC 55484. Bibcode: 2001PNAS...98.9517Y. https://works.bepress.com/edward_yu/3/download/.
- ↑ "Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors". Proc. Natl. Acad. Sci. U.S.A. 104 (8): 2885–90. February 2007. doi:10.1073/pnas.0609359104. PMID 17299051.
 |

