Biology:Tyrosylprotein sulfotransferase
| Tyrosylprotein sulfotransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
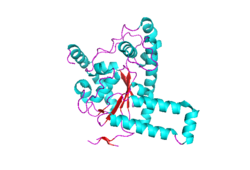 An image of a single subunit of the catalytic region of TPST-2 from protein structure 3AP1 | |||||||||
| Identifiers | |||||||||
| EC number | 2.8.2.20 | ||||||||
| CAS number | 87588-33-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Tyrosylprotein sulfotransferase is an enzyme that catalyzes tyrosine sulfation.[1]
Function
Tyrosylprotein sulfotransferase is the enzyme that catalyzes the sulfation reaction of protein tyrosines, a post-translational modification of proteins. It utilizes 3'-Phosphoadenosine-5'-phosphosulfate (PAPS) as the sulfonate donor and binds proteins with target tyrosine residues to eventually form the tyrosine O-sulfate ester group and the desulfonated 3’-phosphoadenosine-5’-phosphate (PAP).[2][3][4]
TPST and tyrosine sulfation is involved in a large number of biological and physiological processes. Tyrosine sulfation has been found to be an important part of the inflammatory process, leukocyte movement and cytosis, viral cell entrance, and other cell-cell and protein-protein interactions.[2][3] Selection for specific tyrosine residues requires a generally accessible tyrosine residue, and acidic residues within +5 or -5 residues of the target tyrosine.[2][3][4] P-selectin glycoprotein ligand-1 (PSGL-1) has been extensively studied as a substrate for TPST and the importance of sulfation in PSGL-1 and its ability to bind its receptor.[5] Another substrate for TPST, CC-chemokine Receptor 5 (CCR5), has generated interest because of its role as the target protein for the viral entrance of HIV into cells. The importance of CCR5's sulfation for HIV invasion has led to research on TPST and CCR5, including a characterization of the pattern of sulfation of CCR5.[6] Beyond these two proteins, other notable protein substrates include Cholecystokinin (CCK), Factor V and Factor VIII, gastrin, the leech enzyme hirudin, fibrinogen, Complement component 4, follicle-stimulating hormone receptor (FSHR), and other chemokine and G-protein coupled receptors.[2][3] A full, up-to-date list can be found at UniProtKB.
Characterization and properties
Tyrosylprotein sulfotransferase (TPST) is a type II transmembrane protein.[7] It consists of a short cytosolic region that contains the N-terminus of the protein, a single transmembrane region of about 17 amino acids in length, a small stem region of about 40 amino acids in length, and a larger, catalytic region that is located on the luminal side of the membrane.[2][4] It is localized to the Golgi apparatus, specifically in the trans-Golgi region, and acts almost exclusively on secretory and plasma membrane proteins.[8] TPST is about 50-54 kD in size, and has two confirmed isoforms in mammals, TPST-1 and TPST-2, that are 370 and 377 residues in length, respectively.[7][9] Both are quite similar with an approximately 63% amino acid identity, but show slightly different protein substrate specificities.[2][4]
TPST is a prevalent enzyme, found in many multicellular eukaryotes including mammals, most vertebrates, and a number of invertebrate species as well, including Drosophila melanogaster.[2][3][10] Its importance can be further demonstrated by the fact as much as 1% of all secreted and membrane tyrosine residues are found to be sulfated.[6][11]
Mechanism
Within the last two years, using the crystallized structure of the catalytic region of TPST-2 and different experiments other methods using mass spectrometry methods have come to propose two separate mechanisms.
Two-site ping-pong mechanism
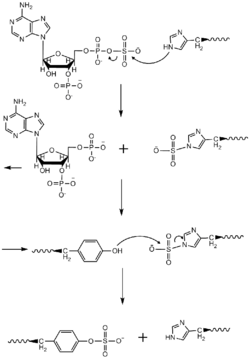
A two-site ping-pong mechanism for TPST and the tyrosine sulfating has been proposed. PAPS enters one site of TPST and the sulfonate group is transferred to a Histidine residue in the enzyme and PAP is released. Then, the target protein and tyrosine bind TPST and the histidine transfers the sulfonate group to the target tyrosine.[11]
SN2-like in-line displacement mechanism
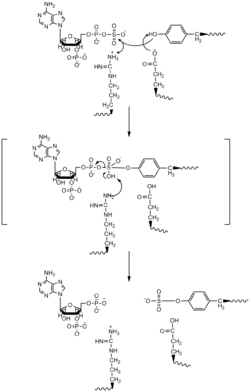
Based on crystal structure of TPST-2 with C4 complement and PAP, an SN2-like in-line displacement mechanism has been proposed. In this mechanism, both PAPS and the target tyrosine bind to the same active site in the enzyme and are orientated in a way such that a glutamic acid residue acts as a catalytic base on the tyrosine hydroxyl group, an arginine residue acts as a catalytic acid, and serine and lysine residues are used to stabilize the SN2-like intermediate. The deprotonated hydroxyl would attack the sulfonate group, then displace the phosphate group and PAP would be released, along with the sulfotyrosine residue.[4]
Examples
Human genes that encode protein-tyrosine sulfotransferase enzymes include:
|
| ||||||||||||||||||||||||||||||||||||||||||||
See also
References
- ↑ "Tyrosine-O-sulfated proteins of PC12 pheochromocytoma cells and their sulfation by a tyrosylprotein sulfotransferase". J. Biol. Chem. 258 (18): 11326–34. September 1983. doi:10.1016/S0021-9258(17)44421-8. PMID 6577005.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 "Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins.". New Biotechnology 25 (5): 299–317. Jun 2009. doi:10.1016/j.nbt.2009.03.011. PMID 19658209.
- ↑ 3.0 3.1 3.2 3.3 3.4 "Protein tyrosine sulfation, 1993--an update.". Chemico-Biological Interactions 92 (1–3): 257–71. Jun 1994. doi:10.1016/0009-2797(94)90068-x. PMID 8033259.
- ↑ 4.0 4.1 4.2 4.3 4.4 "Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.". Nature Communications 4: 1572. 2013. doi:10.1038/ncomms2593. PMID 23481380. Bibcode: 2013NatCo...4.1572T.
- ↑ "Tyrosine sulfation: a modulator of extracellular protein-protein interactions.". Chemistry & Biology 7 (3): R57-61. Mar 2000. doi:10.1016/s1074-5521(00)00093-4. PMID 10712936.
- ↑ 6.0 6.1 "Tyrosine sulfation of CCR5 N-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence.". Proceedings of the National Academy of Sciences of the United States of America 99 (17): 11031–6. Aug 20, 2002. doi:10.1073/pnas.172380899. PMID 12169668. Bibcode: 2002PNAS...9911031S.
- ↑ 7.0 7.1 "Tyrosylprotein sulfotransferase: purification and molecular cloning of an enzyme that catalyzes tyrosine O-sulfation, a common posttranslational modification of eukaryotic proteins.". Proceedings of the National Academy of Sciences of the United States of America 95 (6): 2896–901. Mar 17, 1998. doi:10.1073/pnas.95.6.2896. PMID 9501187. Bibcode: 1998PNAS...95.2896O.
- ↑ "(Glu62, Ala30, Tyr8) n serves as high-affinity substrate for tyrosylprotein sulfotransferase: a Golgi enzyme.". Proceedings of the National Academy of Sciences of the United States of America 82 (18): 6143–7. Sep 1985. doi:10.1073/pnas.82.18.6143. PMID 3862121. Bibcode: 1985PNAS...82.6143L.
- ↑ "Existence of distinct tyrosylprotein sulfotransferase genes: molecular characterization of tyrosylprotein sulfotransferase-2.". Proceedings of the National Academy of Sciences of the United States of America 95 (19): 11134–9. Sep 15, 1998. doi:10.1073/pnas.95.19.11134. PMID 9736702. Bibcode: 1998PNAS...9511134B.
- ↑ "Fluorescence assay for protein post-translational tyrosine sulfation.". Analytical and Bioanalytical Chemistry 405 (4): 1425–9. Feb 2013. doi:10.1007/s00216-012-6540-3. PMID 23161068.
- ↑ 11.0 11.1 "Catalytic mechanism of Golgi-resident human tyrosylprotein sulfotransferase-2: a mass spectrometry approach.". Journal of the American Society for Mass Spectrometry 21 (9): 1633–42. Sep 2010. doi:10.1016/j.jasms.2010.03.037. PMID 20462768.
External links
- tyrosylprotein+sulfotransferase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |