Chemistry:Chloroeremomycin
| |
| Names | |
|---|---|
| Other names
Chloroorienticin A, A82846B, LY264826
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| KEGG | |
PubChem CID
|
|
| UNII | |
| |
| |
| Properties | |
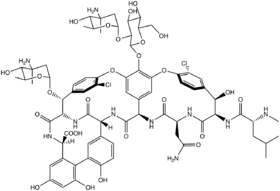
| C73H88Cl2N10O26 | |
| Molar mass | 1592.45 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Chloroeremomycin is a member of the glycopeptide family of antibiotics, such as vancomycin.[1] The molecule is a non-ribosomal polypeptide that has been glycosylated. It is composed of seven amino acids and three saccharide units. Although chloroeremomycin has never been in clinical phases, oritavancin, a semi-synthetic derivative of chloroeremomycin, has been investigated.
Chloroeremomycin is a type of glycopeptide antibiotic and works by blocking the construction of a cell wall. Chloroeremomycin is naturally produced by Amycolatopsis orientalis.
History
Chloroeremomycin was discovered by Eli Lilly in the 1980s. In the 1990s, researchers at Eli Lilly developed biphenyl-chloroeremomycin, now known as oritavancin, as a functionalized derivative of chloroeremomycin to combat rising antibacterial resistance to vancomycin.[2] The chloroeremomycin gene cluster was sequenced by van Wageningen et al in 1998.[3] After the publication, many groups expressed the genes and conducted experiments to understand how chloroeremomycin and, by extension, vancomycin are biosynthesized.[citation needed]
Structure
Chloroeremomycin is composed of seven amino acids (three non-proteinogenic, and four proteinogenic) and three saccharide units. From N-terminus to C-terminus, the order is: Me-L-Leu, L-Tyr, D-Asn, D-4-hydroxyphenylglycine (HPG), L-HPG, D-Tyr, and D-3,5-dihydroxyphenylglycine (DHPG). When referring to specific amino acids, this article will reference the amino acid in the order it appears within the heptapeptide. Chloroeremomycin is glycosylated at aa4 with a Glc-(2→α1)-epivancosamine disaccharide and at aa6 with a D-BHT-(→α1)-epivancosamine saccharide.[citation needed]
Some amino acids are modified prior to the completion of the heptapeptide (in cis) and some are modified after the heptapeptide is formed (in trans). During the synthesis of the heptapeptide, the stereocenters of aa3, aa4, aa6, and aa7 are changed from L to D. Both Tyr residues are hydroxylated and chlorinated after the amino acids have been incorporated to the growing polypeptide to form 4-chloro-β-hydroxytyrosine (BHT). The now-BHT residues are then crosslinked to the aa4 HPG through aryl-ether linkages. An aryl-aryl bond is formed between aa5 and aa7 at the aa5-C3 and aa7-C2 positions on the aromatic rings. Finally, the N-terminus Leu is methylated.[citation needed]
In addition to the presence of D-amino acids, the molecule has atropisomer chemistry. The orientations of the chloro-substituted phenyl rings add another aspect of stereochemistry to the molecule.
Biosynthesis
Chloroeremomycin was found to be synthesized by Amycolatopsis orientalis.
Non-ribosomal peptide synthase
The non-ribosomal peptide synthase (NRPS) is encoded by three genes: CepA, CepB, and CepC. CepA links the first three amino acids; CepB adds the fourth to sixth amino acids; CepC adds the last amino acid and includes a thioesterase domain to release the heptapeptide from the NRPS complex.[4] The growing peptide chain is passed through modules for each amino acid. The basic organization of each module is A-PCP-C. The A, or adenylation, region activates the domain's amino acid to allow transfer to the PCP, or peptide carrying protein, region. The activated amino acid is transferred to a cysteine residue in the PCP region, which anchors the amino acid and prepares the amino acid to be added to the polypeptide. The C, or condensation, region attaches the amino acid to the polypeptide. In addition, modules 2, 4, and 5 have E regions that epimerize (switch the stereochemistry) of the added amino acid to produce the correct configuration. Module 7, the last module, has an X and TE region. The X region is responsible for recruiting several of the tailoring enzymes that will perform the necessary reactions (halogenation, glycosylation, methylation, oxidative cross-linking, and hydroxylations) to produce chloroeremomycin.[5] Finally, the TE, or thioesterase, region releases chloroeremomycin from the NRPS complex.
Post-peptide modifications
The modification required to biosynthesize mature chloroeremomycin include: oxidative cross-linking of aromatic rings, hydroxylation and chlorination of the two Tyr residues, methylation of Leu, and glycosylation at aa4 and aa6.[3]
The oxidative crosslinks are catalyzed by enzymes OxyA-C. The glycosylations are catalyzed by enzymes GtfA-C (coded by Orf11-13 respectively). The chlorinations are performed by enzymes encoded by Orf10 and 18.[citation needed]
Total synthesis
There is no reported total synthesis of chloroeremomycin, although there are several total syntheses of vancomycin. The structures of vancomycin and chloroeremomycin are very similar, differing only in the glycosylation sites. Vancomycin is glycosylated at aa4 with a (2-beta1)-Glc-vancosamine disaccharide. As mentioned above, chloroeremomycin is glycosylated at aa4 with a (2-beta1)-Glc-epivancosamine disaccharide and at aa6 with a beta1-epivancosamine saccharide.
Pharmacology and chemistry
See also
- Glycorandomization
- Teixobactin
References
- ↑ Wright, Andrew G. McArthur, Gerard D.. "The Comprehensive Antibiotic Resistance Database". https://card.mcmaster.ca/ontology/36323.
- ↑ Sharman, G. J., Try, A. C., Dancer, R. J., Cho, Y. R., Staroske, T., Bardsley, B., Maguire, A. J., Cooper, M. A., O'Brien, D. P., Williams, D. H. "The Roles of Dimerization and Membrane Anchoring in Activity of Glycopeptide Antibiotics against Vancomycin-Resistant Bacteria." J. Am. Chem. Soc. 1997, 119, 12041-12047.
- ↑ 3.0 3.1 van Wageningen, A. M. A., Kirkpatrick, P. N., Williams, D. H., Harris, B. R., Kershaw, J. K., Lennard, N. J., Jones, M., Jones, S. J. M., Solenberg, P. J. "Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic." Chemistry & Biology, 1997, 5, 155-162.
- ↑ Yim, G., Thaker, M. N., Koteva, K., Wright, G. "Glycopeptide antibiotic biosynthesis." The Journal of Antibiotics, 2017, 67, 31-41.
- ↑ Haslinger, K., Peschke, M., Brieke, C., Maximowitsch. E., Cryle, M. J. "X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis." Nature, 2015, 521, 105-110.
 |

