History:Denisovan
The Denisovans or Denisova hominins (/dəˈniːsəvə/ də-NEE-sə-və) are an extinct species or subspecies of archaic human that ranged across Asia during the Middle to Late Pleistocene, approximately 200,000–40,000 years ago. Most of what is known about Denisovans comes from DNA evidence. While many recent fossils have been found and tentatively identified as Denisovan, the first Denisovans discovered were known from few physical remains. Consequently, no formal species name has been established. However, an analysis of the mitochondrial DNA and endogenous proteins from the Harbin cranium, which had been given the name Homo longi, showed with great certainty that this species represents a Denisovan.[1][2][3] In a study published in September 2025, remains from six additional sites in China including the 1 million year old Yunxian man were proposed to be included in the species Homo longi along with the genetically confirmed Denisovans.[4]
The first identification of a Denisovan individual occurred in 2010, based on mitochondrial DNA (mtDNA) extracted from a juvenile finger bone excavated from the Siberian Denisova Cave in the Altai Mountains in 2008.[5][6] Nuclear DNA indicates close affinities with Neanderthals. The cave was also periodically inhabited by Neanderthals. Additional specimens from Denisova Cave were subsequently identified, as were specimens from the Baishiya Karst Cave on the Tibetan Plateau, Tam Ngu Hao 2 Cave in the Annamite Mountains of Laos, the Penghu channel between Taiwan and the mainland, and Harbin in Manchuria.
DNA evidence suggests they had dark skin, eyes, and hair, and had a Neanderthal-like build.[7][8] Based on the Harbin cranium, like other archaic humans, the skull is low and long, with massively developed brow ridges, wide eye sockets, and a large mouth. The two existing Denisovan mandibles show that like Neanderthals, the Denisovans lacked a chin. Like modern humans and the much earlier Homo antecessor, but unlike Neanderthals, the face is rather flat, but with a larger nose. However, they had larger molars which are reminiscent of Middle to Late Pleistocene archaic humans and australopithecines. The cranial capacity and therefore the brain size of the Denisovans was within the range of modern humans and Neanderthals.
Denisovans interbred with modern humans, with a high percentage (roughly 5%) of Denisovan DNA occurring in Melanesians, Aboriginal Australians, and Filipino Negritos.[9] In contrast, 0.2% derives from Denisovan ancestry in mainland Asians and Native Americans.[10] In a 2018 study, South Asians were found to have levels of Denisovan admixture similar to that seen in East Asians.[11] Another study found that the highest Denisovan ancestry is inferred in Oceanians (~2.0%), while most populations of Native Americans, East Asians, and South Asians have similar amounts (~0.1%).[12] This distribution suggests that there were Denisovan populations across Asia. There is also evidence of interbreeding with the Altai Neanderthal population, with about 17% of the Denisovan genome from Denisova Cave deriving from them. A first-generation hybrid nicknamed "Denny" was discovered with a Denisovan father and a Neanderthal mother.[13][14] Additionally, 4% of the Denisovan genome comes from an unknown archaic human species, which diverged from modern humans over one million years ago.[15]
Taxonomy
|
Cladogram based on palaeoproteomics:[1][16][17]
Cladogram of Homo longi (Denisovans) based on Feng et al., 2025:[4]
|
Denisovans might represent a new species of Homo or an archaic subspecies of Homo sapiens (modern humans), but up until the Harbin cranium was identified as a Denisovan in June 2025 through the mitochondrial DNA[2] and autosomal proteomics[1] there were too few fossils to erect a proper taxon. Proactively proposed species names without type specimen designation for Denisovans include H. sapiens altaiensis or H. altaiensis (Derevianko, 2011),[18] H. denisoviensis (Picq, 2011),[19] H. denisovan (Gabriel & Mihaela, 2011),[20] and H. denisova (Gunbin et al., 2012).[21]
In 2025, Denné Reed argued that the informal name "Denisovans" represents the better system than proactively proposed names to reference this archaic human group due to its uncertain biological status as an independent evolutionary lineage. He suggested that "H. altaiensis" represents a nomen nudum ("naked name"), since its description lacks differential diagnosis, does not clearly display the intent of naming a new species and lacks a fixed type specimen. He also suggested that the names "Homo daliensis" and "Homo mapaensis" are conditionally proposed which makes them unavailable based on ICZN article 15, while considered "Homo tsaichangensis" (intended name for Penghu 1 in a self-published digital book) to be unpublished and unavailable, as it does not contain evidence of ZooBank registration within the published work which fails to conform to the ICZN articles 8.5.3.1 and 8.5.3.2.[22]
Research published in 2024 proposed classifying Denisovans as part of the conditional species Homo juluensis based on the similarities between Denisovan and H. juluensis molars, prior to the classification of Denisovans as Homo longi based on DNA evidence.[23][24]
Some older findings called "East Asian Archaics" have been associated in studies with the Denisovans but may or may not belong to the Denisovan line. Such findings include the Dali skull,[25] the Xujiayao hominin,[26] the Xuchang crania,[27] the Jinniushan human, the Hualongdong people,[28] Yunxian Man,[29] Maba Man,[30] and the Narmada Human.[31]
In 2021, Chinese palaeoanthropologist Qiang Ji and colleagues suggested that their newly erected species, H. longi, may represent the Denisovans, based on the similarity between the type specimen's molar and that of the Xiahe mandible.[32] In 2024, paleoanthropologists Christopher Bae and Xiujie Wu designated the Xujiayao fossils as the holotype of the species Homo juluensis with Xuchang as the paratype, and suggested sinking Denisovans into this species. They recommended relegating the Dali Man and the similar specimen Jinniushan to H. longi.[33][34]
In 2025, Fu and colleagues retrieved mitochondrial DNA from the dental calculus of the Harbin cranium (H. longi holotype), reporting that it falls within the variation of seven previously sequenced Denisovan mitochondrial DNA.[2] Fu et al. (2025) also retrieved 95 endogenous proteins from the same specimen, and suggested that H. longi can be confidently assigned to a Denisovan population.[1]
In 2025, Feng and colleagues, the team that published the Harbin cranium in 2021 and named the new species Homo longi,[32] ran a morphometric analysis of 104 ancient hominin cranial and mandibular specimens using 533 morphometric landmarks and grouped Yunxian Man, Dali Man, the Hualongdong people, the Jinniushan human, the Xujiayao hominins, and Maba Man under H. longi alongside the genetically confirmed Denisovans.[4]
Discovery

The Denisova Cave is located in Altai Krai, Russia, in south-central Siberia, on the western edges of the Altai Mountains. It is named after Denis (Dyonisiy), a Russian Old Believer hermit who lived there in the 18th century. The cave was first inspected for fossils in the 1970s by Soviet paleontologist Nikolai Ovodov, who was looking for remains of canids.[35]
In 2008, Michael Shunkov from the Russian Academy of Sciences and other Russian archaeologists from the Institute of Archaeology and Ethnography of the Siberian Branch of the Russian Academy of Sciences in Novosibirsk Akademgorodok investigated the cave and found the finger bone of a juvenile female hominin originally dated to 50–30,000 years ago.[5][36] The estimate has changed to 76,200–51,600 years ago.[37] The specimen was originally named X-woman because matrilineal mitochondrial DNA (mtDNA) extracted from the bone demonstrated it to belong to a novel ancient hominin, genetically distinct both from contemporary modern humans and from Neanderthals.[5]
In 2019, Greek archaeologist Katerina Douka and colleagues radiocarbon dated specimens from Denisova Cave, and estimated that Denisova 2 (the oldest specimen) lived 195,000–122,700 years ago.[37] Older Denisovan DNA collected from sediments in the East Chamber dates to 217,000 years ago. Based on artifacts also discovered in the cave, hominin occupation (most likely by Denisovans) began 287±41 or 203±14 ka. Neanderthals were also present 193±12 ka and 97±11 ka, possibly concurrently with Denisovans.[38]
Specimens
The fossils of multiple distinct Denisovan individuals from Denisova Cave have been identified through their ancient DNA (aDNA): Denisova 2, 3, 4, 8, 11, 19, 20, 21 and 25.[39] An mtDNA-based phylogenetic analysis of these individuals suggested that Denisova 19, 20, and 21 are the oldest, followed by Denisova 2, then Denisova 8; while Denisova 3 and Denisova 4 were roughly contemporaneous.[40][39] The mtDNA from Denisova 4 bore a high similarity to that of Denisova 3, indicating that they belonged to the same population.[41]
Denisova Cave contained the only known examples of Denisovans until 2019, when a research group led by Fahu Chen, Dongju Zhang, and Jean-Jacques Hublin described a partial mandible discovered in 1980 by a Buddhist monk in the Baishiya Karst Cave on the Tibetan Plateau in China. Known as the Xiahe mandible, the fossil became part of the collection of Lanzhou University, where it remained unstudied until 2010.[42] It was determined by ancient protein analysis to contain collagen that by sequence was found to have close affiliation to that of the Denisovans from Denisova Cave, while uranium decay dating of the carbonate crust enshrouding the specimen indicated it was more than 160,000 years old.[43] The identity of this population was later confirmed through study of environmental DNA, which found Denisovan mtDNA in sediment layers ranging in date from 100,000 to 60,000 years before present, and perhaps more recent.[44] A 2024 reanalysis identified a partial Denisovan rib fragment dating to between 48,000 and 32,000 BP.[45][46]
In 2018, a team of Laotian, French, and American anthropologists, who had been excavating caves in the Laotian Annamite Mountains since 2008, was directed by local children to the site Tam Ngu Hao 2 ("Cobra Cave") where they recovered a human tooth. The tooth (catalogue number TNH2-1) developmentally matches a 3.5 to 8.5 year old, and a lack of amelogenin (a protein on the Y chromosome) suggests it belonged to a girl, barring extreme degradation of the protein over a long period of time. Dental proteome analysis was inconclusive for this specimen, but the team found it anatomically comparable with the Xiahe mandible, and so they categorized it as a Denisovan. The tooth probably dates to 164,000 to 131,000 years ago.[47]
In 2022, a team from Germany, Austria, Russia and the UK found three Denisovans (Denisova 19, 20, 21) from layer 15 of the East Chamber, in Denisova Cave. It turns out that the mtDNA sequences of Denisova 19 and 21 are identical, indicating that they may belong to the same individual or be maternal relatives. The divergence date for the mtDNAs of the three new and the four previously published Denisovans is 229 ka (206–252 ka 95% Cl) during the Interglacial period MIS 7.[39] The genetic diversity among the Denisovans from Denisova Cave is on the lower range of what is seen in modern humans, and is comparable to that of Neanderthals. However, it is possible that the inhabitants of Denisova Cave were more or less reproductively isolated from other Denisovans, and that, across their entire range, Denisovan genetic diversity may have been much higher.[40]
In 2024, scientists announced the sequence of Denisova 25, which was in a layer dated to 200 ka.[48] During DNA sequencing, low proportions of the Denisova 2, Denisova 4 and Denisova 8 genomes were found to have survived, but high proportions of the Denisova 3 and Denisova 25 genomes were intact.[40][49][48] The Denisova 3 sample was cut into two, and the initial DNA sequencing of one fragment was later independently confirmed by sequencing the mtDNA from the second.[50]
In 2008, a Taiwanese citizen purchased a fossil Homo mandible, dredged from the sea floor of the Taiwan Strait, from an antique shop and donated it to the Taiwan National Museum of Natural Science. Attempts to extract DNA were unsuccessful, but in 2025 protein analysis of the specimen, designated Penghu 1, was published showing that it belonged to a male Denisovan.[51]
In 2018, a relatively complete skull was reported from Harbin, China, and was described in 2021 as H. longi.[32] In 2025, mtDNA and proteomic analysis confirmed that this skull is a Denisovan.[2][1]
The following list is the currently known specimens of Denisovans, with colored specimens being proposed through morphometric analyses only without mitogenome or proteomic analyses:[4]
| Name | Fossil elements | Age | Discovery | Place | Sex and age | Publication | Image | GenBank / Genebase accession |
|---|---|---|---|---|---|---|---|---|
| Denisova 3 (also known as X Woman)[41][50][5] |
Distal phalanx of the fifth finger | 76.2–51.6 ka[37] | 2008 | Denisova cave (Russia) | 13.5-year-old adolescent female | 2010 |  |
NC013993 |
| Denisova 4[41][25] | Permanent upper 2nd or 3rd molar | 84.1–55.2 ka[37] | 2000 | Denisova cave (Russia) | Adult male | 2010 | 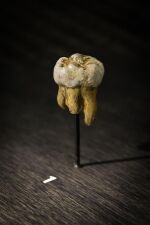 |
FR695060 |
| Denisova 8[49] | Permanent upper 3rd molar | 136.4–105.6 ka[37] | 2010 | Denisova cave (Russia) | Adult male | 2015 | KT780370 | |
| Deciduous 2nd lower molar | 194.4–122.7 ka[37] | 1984 | Denisova cave (Russia) | Adolescent female | 2017 | KX663333 | ||
| Xiahe mandible[43] | Partial mandible | > 160 ka | 1980 | Baishiya Cave (China) | 2019 | 150px | ||
| Penghu 1[52][51] | Partial mandible | 130 to 190 kya or 10 to 70 kya | 2008 | Penghu Channel (Taiwan) | Adult male | 2015 |  |
|
| Arm or leg bone fragment | 118.1–79.3 ka[37] | 2012 | Denisova cave (Russia) | 13 year old adolescent female | 2016 | 150px | ||
| Denisova 13[53] | Parietal bone fragment | Found in layer 17 (202-167 ka)[54] | 2019 | Denisova cave (Russia) | pending | |||
| TNH2–1[47] | Permanent lower left 1st or 2nd molar | 164–131 ka | 2018 | Tam Ngu Hao 2 cave (Laos) | 3.5 to 8.5 year old female | 2022 | 150px | |
| Denisova 19[39] | Undiagnostic bone fragment | Layer 15, 217–187 ka | 2012–13 | Denisova cave (Russia) | 2022 | |||
| Denisova 20[39] | Undiagnostic bone fragment | Layer 15, 217–187 ka | 2012–13 | Denisova cave (Russia) | 2022 | |||
| Denisova 21[39] | Undiagnostic bone fragment | Layer 15, 217–187 ka | 2012–13 | Denisova cave (Russia) | 2022 | |||
| Distal rib fragment | 42 ka | 1980 | Baishiya Cave (China) | Unknown | 2024 | 150px | ||
| Denisova 25[48] | Molar | 200 ka | 2024 | Denisova cave (Russia) | Male | pending | ||
| Harbin cranium (holotype of Homo longi, also known as the Dragon Man)[6][32] | Complete skull | >146 ka | 1933?[55] | Harbin (China) | Adult male aged approximately 50 years[56] | 2021 | 150px | C_AA108316 |
| Dali cranium[57] | Complete skull | 260±20 ka[58] | 1978 | Dali County (China) | Adult | 1981 | 150px | |
| Jinniushan human[59][60] | Fragments of the skull cap (cranium), ribs, hand, pelvis, and leg bones | 260±20 ka | 1984 | Jinniushan (China) | Adult female[61] | 1985 | 150px | |
| Xujiayao hominin[62][63] | 12 parietal bones, 1 temporal bone, 2 occipital bones, 1 mandibular bone fragment, 1 juvenile maxilla, and 3 isolated teeth. | 130–71 ka[64] | 1976–1979 | Xujiayao village in Yanggao County (China) | Adult and juvenile | 2011 | 150px | |
| HLD 6[65][66] | 11 fossil parts belong to a single individual | 300 ka | 2014–2016, 2020 | Hualong Cave (China) | Juvenile 12–13 years old[66] | 2019 | ||
| Maba 1 (PA 84)[67][68] | Partial skull, a skull cap and parts of the right upper face, with parts of the nose also still attached | 300–130 ka[69][67] | 1958 | Lion Cave (Shiziyan), Maba, near Shaoguan city in the northern part of Guangdong province (China) | Adult middle-aged male[67] | 1959 | 150px |
Evolution
The nuclear DNA (nDNA) of Denisova 3—which had an unusual degree of DNA preservation with only low-level contamination— shows that Denisovans and Neanderthals were more closely related to each other than they were to modern humans. Using the percent distance from human–chimpanzee last common ancestor, Denisovans/Neanderthals split from modern humans about 804,000 years ago, and from each other 640,000 years ago.[41] Using a mutation rate of 1×10−9 or 0.5×10−9 per base pair (bp) per year, the Neanderthal/Denisovan split occurred around either 236–190,000 or 473–381,000 years ago respectively.[70] Using 1.1×10−8 per generation with a new generation every 29 years, the time is 744,000 years ago. Using 5×10−10 nucleotide site per year, it is 616,000 years ago. Using the latter dates, the split had likely already occurred by the time hominins spread out across Europe.[71] H. heidelbergensis is typically considered to have been the direct ancestor of Denisovans and Neanderthals, and sometimes also modern humans.[72] Due to the strong divergence in dental anatomy, they may have split before characteristic Neanderthal dentition evolved about 300,000 years ago.[41]

Sequenced mitochondrial DNA (mtDNA), preserved by the cool climate of the cave (average temperature is at freezing point), was extracted from Denisova 3 by a team of scientists led by Johannes Krause and Svante Pääbo from the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany. Denisova 3's mtDNA differs from that of modern humans by 385 bases (nucleotides) out of approximately 16,500, whereas the difference between modern humans and Neanderthals is around 202 bases. In comparison, the difference between chimpanzees and modern humans is approximately 1,462 mtDNA base pairs. This suggested that Denisovan mtDNA diverged from that of modern humans and Neanderthals about 1,313,500–779,300 years ago; whereas modern human and Neanderthal mtDNA diverged 618,000–321,200 years ago.[5][73]
Modern humans contributed mtDNA to the Neanderthal lineage, but not to the Denisovan mitochondrial genomes yet sequenced.[74][75][76][77] The mtDNA sequence from the femur of a 400,000-year-old early Neanderthal from the Sima de los Huesos Cave in Spain was found to be closer to Denisovans,[2][78][79] and the authors posited that this mtDNA represents an archaic sequence which was subsequently lost in Neanderthals due to replacement by a modern-human-related sequence.[80]
In 2020, the sequencing of Denisovan Y chromosomes (Denisova 4 and Denisova 8), as well as the Y chromosomes of three late Neanderthals (Spy 94a, Mezmaiskaya 2 and El Sidrón 1253) showed that the Denisovan Y chromosomes split around 700 thousand years ago (kya) from a lineage shared by Neanderthal and modern human Y chromosomes, which diverged from each other around 370 thousand years ago. The phylogenetic relationships of archaic and modern human Y chromosomes differ from the population relationships inferred from the autosomal genomes and mirror mitochondrial DNA phylogenies, indicating replacement of both the mitochondrial and Y chromosomal gene pools in late Neanderthals.[81]
Demographics

Denisovans are known to have lived in Siberia, Tibet, Laos, Taiwan and Manchuria.[47][51][1][2] The Xiahe mandible is the earliest recorded human presence on the Tibetan Plateau.[43] Though their remains have been identified in only these five locations, traces of Denisovan DNA in modern humans suggest they ranged across East Asia,[82][83]
In 2019, geneticist Guy Jacobs and colleagues identified three distinct populations of Denisovans responsible for the introgression into modern populations now native to, respectively: Siberia and East Asia; New Guinea and nearby islands; and Oceania and, to a lesser extent, across Asia. Using coalescent modeling, the Denisova Cave Denisovans split from the second population about 283,000 years ago; and from the third population about 363,000 years ago. This indicates that there was considerable reproductive isolation between Denisovan populations.[84] Using exponential distribution analysis on haplotype lengths, Jacobs calculated introgression into modern humans occurred about 29,900 years ago with the Denisovan population ancestral to New Guineans; and 45,700 years ago with the population ancestral to both New Guineans and Oceanians. A third wave appears to have introgressed into East Asia, but there is not enough DNA evidence to pinpoint a solid timeframe.[84]
In a 2024 study, scientist Danat Yermakovich, of the University of Tartu, discovered that people living at different elevations in Papua New Guinea have differences in Denisovan DNA; with people living in the highlands having variants for early brain development and those living in the lowlands having variants for the immune system.[85] Based on the high percentages of Denisovan DNA in modern Papuans and Australians, Denisovans may have crossed the Wallace Line into these regions (with little back-migration west), the second known human species to do so,[31] along with earlier Homo floresiensis. By this logic, they may have also entered the Philippines, living alongside H. luzonensis which, if this is the case, may represent the same or a closely related species.[86] These Denisovans may have needed to cross large bodies of water.[84] Alternately, high Denisovan DNA admixture in modern Papuan populations may simply represent higher mixing among the original ancestors of Papuans prior to crossing the Wallace line.
Denisova Cave, over time of habitation, continually swung from a fairly warm and moderately humid pine and birch forest to tundra or forest-tundra landscape.[38] Conversely, Baishiya Karst Cave is situated at a high elevation, an area characterized by low temperature, low oxygen, and poor resource availability. Colonization of high-altitude regions, due to such harsh conditions, was previously assumed to have only been accomplished by modern humans.[43] Denisovans seem to have also inhabited Southeast Asia.[83] The Tam Ngu Hao 2 site might have been a closed forest environment.[47]
Anatomy
The finger bone is within the modern human range of variation for women,[50] which is in contrast to the large, robust molars which are more similar to those of Middle to Late Pleistocene archaic humans. All the available molars are outside the range of any Homo species except H. habilis and H. rudolfensis, and is more like those of australopithecines. The second molar is larger than those of modern humans and Neanderthals, and is more similar to those of H. erectus and H. habilis.[41] Like Neanderthals, the mandible had a gap behind the molars, and the front teeth were flattened; but Denisovans lacked a high mandibular body, and the mandibular symphysis at the midline of the jaw was more receding.[43][87]
The Denisovan Harbin cranium, Homo longi is characterized by a low and long skull, receding forehead, extremely wide upper face, a large nasal opening equating to an enlarged nose (possibly an adaptation to the cold air), large and square eye sockets, inflated and thick brow ridges (supraorbital torus), flat cheekbones (zygomatic bone), a wide palate and large tooth sockets (equating to a large mouth), and a broad base of the skull.[32] The Harbin skull is the longest archaic human skull to date. The Harbin skull also has the longest brow ridge of any archaic or modern cranium.[56]

The Harbin cranium had a massive brain at roughly 1,420 cc, above the range of all known human species except modern humans and Neanderthals. Nonetheless, post-orbital constriction (a constriction of the braincase just behind the eyes, absent in modern humans, and equating to the location of the frontal lobes) is more developed in H. longi than in Neanderthals, although not so much as in more-ancient human species.[32]
Despite the face being so wide, it was rather flat (reduced mid-facial prognathism), and resembles the anatomy found in modern humans, the far more ancient H. antecessor, and other Middle Pleistocene Chinese specimens. This gives the Harbin cranium features that are similar to that of modern humans, but in fact, this is likely an ancestral trait.[89]
Because the original describers judged the Harbin skull to be closely allied with the Xiahe mandible, they believed H. ongi lacked a chin, like other archaic humans, but the specimen's lower jaw was not recovered.[56] This lack of a chin is also present in the Denisovan Penghu mandible.[51]
Both Ni et al. (2021)[56] and Bae and Wu (2024)[88] group the Harbin cranium, Homo longi, together with the Dali cranium and the skeleton from Jinniushan into a Homo longi clade. Feng et al. (2025)[4] group Yunxian Man, Dali Man, the Hualongdong people, the Jinniushan human, the Xujiayao hominins, and Maba Man under H. longi alongside the genetically confirmed Denisovans. Therefore, according to these studies, the morphological characteristics of these remains can be also applied to Denisovans in general.
Because Jinniushan has abundant post-cranial remains, and is also grouped with Homo longi in several studies, it is very likely that these remains also belong to a Denisovan as well. This would enable a description of the post-cranial (body) morphology of the Denisovans.[56][88][4]
The human fossil elements at Jinniushan all belong to one female individual. The fossil remains consist of one cranium, six vertebrae (one cervical, five thoracic), one complete left os coxae (pelvic bone), one complete left ulna, one complete left patella (kneecap), two left ribs, and several hand and feet bones. Analysis of the left-half of the pelvis (hip bone) in 2006 indicated that the individual was a woman. Like the Harbin cranium, a distinguishing feature of the Jinniushan female is its large brain with a brain size (cranial capacity) of 1,330 cm3 (81 cu in).[90][91] Quoting Rosenberg et al. (2006) the "Reconstructed stature using this formula is 168.78 ± 4.30 cm." "we have used the mean of these two estimates, 78.6 kg, as the estimated body weight". Therefore we have an estimate of the body proportions, height, and weight of an adult female Denisovan (Homo longi), making it the largest female specimen ever discovered in the fossil record, although within the range of modern human females.[91]
The Denisovan genome from Denisova Cave has variants of genes which, in modern humans, are associated with dark skin, brown hair, and brown eyes.[7] The Denisovan genome also contains a variant region around the EPAS1 gene that in Tibetans assists with adaptation to low oxygen levels at high elevation,[92][43] and in a region containing the WARS2 and TBX15 loci which affect body-fat distribution in the Inuit.[93]
Culture
Denisova Cave
Early Middle Paleolithic stone tools from Denisova Cave included cores, scrapers, denticulate tools, and notched tools, deposited about 287±41 thousand years ago in the Main Chamber of the cave; and about 269±97 thousand years ago in the South Chamber; up to 170±19 thousand and 187±14 thousand years ago in the Main and East Chambers, respectively.[38]
Middle Paleolithic assemblages were dominated by flat, discoidal, and Levallois cores, and there were some isolated sub-prismatic cores. There were predominantly side scrapers (a scraper with only the sides used to scrape), but also notched-denticulate tools, end-scrapers (a scraper with only the ends used to scrape), burins, chisel-like tools, and truncated flakes. These dated to 156±15 thousand years ago in the Main Chamber, 58±6 thousand years ago in the East Chamber, and 136±26–47±8 thousand years ago in the South Chamber.[38]
Early Upper Paleolithic artefacts date to 44±5 thousand years ago in the Main Chamber, 63±6 thousand years ago in the East Chamber, and 47±8 thousand years ago in the South Chamber, though some layers of the East Chamber seem to have been disturbed. There was blade production and Levallois production, but scrapers were again predominant. A well-developed, Upper Paleolithic stone bladelet technology distinct from the previous scrapers began accumulating in the Main Chamber around 36±4 thousand years ago.[38]
In the Upper Paleolithic layers, there were also several bone tools and ornaments: a marble ring, an ivory ring, an ivory pendant, a red deer tooth pendant, an elk tooth pendant, a chloritolite bracelet, and a bone needle. However, Denisovans are only confirmed to have inhabited the cave until 55 ka; the dating of Upper Paleolithic artefacts overlaps with modern human migration into Siberia (though there are no occurrences in the Altai region); and the DNA of the only specimen in the cave dating to the time interval (Denisova 14) is too degraded to confirm species identity, so the attribution of these artefacts is unclear.[94][38]
Tibet

The inhabitants of Baishiya Karst Cave seem to have been extensively processing goat antelopes, cows, deer, horses, and woolly rhinoceros. They were also butchering large carnivores (cave hyena, dog, and big cat), marmots, hare, and eagles. They may have also used these animals' long bones to make bone tools, and additionally there are stone artefacts in each layer excavated.[45]
In 1998, five child hand- and footprint impressions were discovered in a travertine unit near the Quesang hot springs in Tibet; in 2021, they were dated to 226 to 169 thousand years ago using uranium decay dating. This is the oldest evidence of human occupation of the Tibetan Plateau, and since the Xiahe mandible is the oldest human fossil from the region (though younger than the Quesang impressions), these may have been made by Denisovan children. The impressions were printed onto a small panel of space, and there is little overlap between all the prints, so they seem to have been taking care to make new imprints in unused space. If considered art, they are the oldest known examples of rock art. Similar hand stencils and impressions do not appear again in the archeological record until roughly 40,000 years ago.[95]
The footprints comprise four right impressions and one left superimposed on one of the rights. They were probably left by two individuals. The tracks of the individual who superimposed their left onto their right may have scrunched up their toes and wiggled them in the mud, or dug their finger into the toe prints. The footprints average 192.3 mm (7.57 in) long, which roughly equates to a 7- or 8-year-old child by modern human growth rates. There are two sets of handprints (from a left and right hand), which may have been created by an older child unless one of the former two individuals had long fingers. The handprints average 161.1 mm (6.34 in), which roughly equates with a 12 year old modern human child, and the middle finger length agrees with a 17 year old modern human. One of the handprints shows an impression of the forearm, and the individual was wiggling their thumb through the mud.[95]
Yunnan
In 2025, archaeologists discovered an assemblage of 35 wooden implements on the shores of an ancient lake from the site of Gantangqing (甘棠箐) in Yunnan southwestern China, which was found associated with stone tools, antler billets (soft hammers), and cut-marked bones and is dated from ~361,000 to ~250,000 years at a 95% confidence interval, at a time when Denisovans were known to have inhabited the general region.[96][97][47] The wooden implements include digging sticks made of pine and hardwood, hooks for cutting roots, and small, complete, hand-held pointed tools.
"Our results suggest that hominins at Gantangqing made strategic utilization of lakeshore food resources," the researchers wrote in the study. "They made planned visits to the lakeshore and brought with them fabricated tools of selected wood for exploiting underground tubers, rhizomes, or corms. As such, the Gantangqing assemblage shows the likely use of underground storage organs and the importance of plant foods in early hominin diets in a subtropical environment."[98]
Interbreeding
{{Graphical timeline
|
| help=off | link-to=Human timeline
| scale-increment=1.000 | plot-colour=#ffc966 | from=-10.000 | to=-0.000
| title=Hominin timeline
| bar1-from=-10.000 | bar1-to=-2.800 | bar1-text=Hominini | bar1-colour=#ffa500 | bar1-left=0.0 | bar1-nudge-right=-0.3 | bar1-nudge-down=-3.3
| bar2-from=-9.800 | bar2-to=-9.700 | bar2-text=Nakalipithecus | bar2-colour=#ffa500 | bar2-left=0.1 | bar2-nudge-down=0 | bar2-nudge-left=0.5
| bar3-from=-9.000 | bar3-to=-8.900 | bar3-text=Ouranopithecus | bar3-colour=#ffa500 | bar3-left=0.1 | bar3-nudge-down=0 | bar3-nudge-left=0.3
| bar4-from=-7.000 | bar4-to=-6.900 | bar4-text=Sahelanthropus | bar4-colour=#ffa500 | bar4-left=0.1 | bar4-nudge-down=0 | bar4-nudge-left=0.3
| bar5-from=-6.000 | bar5-to=-5.900 | bar5-text=Orrorin | bar5-colour=#ffa500 | bar5-left=0.1 | bar5-nudge-down=0 | bar5-nudge-left=2.0
| bar6-from=-4.400 | bar6-to=-4.300 | bar6-text=Ardipithecus | bar6-colour=#ffa500 | bar6-left=0.1 | bar6-nudge-down=0 | bar6-nudge-left=1.0
| bar7-from=-3.600 | bar7-to=-1.200 | bar7-text=Australopithecus | bar7-colour=#ffa500 | bar7-left=0.0 | bar7-nudge-down=4 | bar7-nudge-left=0 | bar7-nudge-right=0.4
| bar8-from=-2.800 | bar8-to=-1.500 | bar8-text=Homo habilis | bar8-colour=#ffb732 | bar8-left=0.1 | bar8-nudge-right=0.4 | bar8-nudge-down=0.3
| bar9-from=-1.900 | bar9-to=-0.035 | bar9-text=Homo erectus | bar9-colour=#ffc966 | bar9-left=0.2 | bar9-nudge-right=0.1 | bar9-nudge-down=1.0
| bar10-from=-1.500 | bar10-to=-1.200 | bar10-text= | bar10-colour=#ffc966 | bar10-left=0.1 | bar10-right=0.2 | bar10-nudge-right=0.0 | bar10-nudge-down=0.0
| bar11-from=-0.700 | bar11-to=-0.2 | bar11-text=H. heidelbergensis | bar11-colour=#ffeeaa | bar11-left=0.1 | bar11-nudge-right=0.5 | bar11-nudge-down=0.3
| bar12-from=-0.3 | bar12-to=-0.000 | bar12-text=Homo sapiens | bar12-colour=#ffff00 | bar12-left=0.4 | bar12-nudge-right=0.0 | bar12-nudge-down=0.0
| bar13-from=-0.035 | bar13-to=-0.000 | bar13-text= | bar13-colour=#ffeeaa | bar13-left=0.0 | bar13-right=0.1 | bar13-nudge-right=0.0 | bar13-nudge-down=0.0
| bar14-from=-0.040 | bar14-to=-0.000 | bar14-text= | bar14-colour=#ffff00 | bar14-left=0.1 | bar14-right=0.4 | bar14-nudge-right=0.0 | bar14-nudge-down=0.0
| bar15-from=-0.25 | bar15-to=-0.04 | bar15-text=Neanderthals | bar15-colour=#ffeeaa | bar15-left=0.1 | bar15-right=0.4 | bar15-nudge-left=0.2 | bar15-nudge-down=0.2
| note3-at=-10.000 | note3=Earlier apes
| note4-at=-9.000
| note4=Gorilla split
| note5-at=-7.000 | note5=Possibly bipedal
| note8-at=-5.800
| note8=Chimpanzee split
| note10-at=-4.050 | note10=Earliest bipedal
| note12-at=-3.300 | note12=Stone tools
| note14-at=-1.800
| note14=Exit from Africa
| note15-at=-1.500 | note15=[[Biology:Control of fire by early humans#Lower Paleolithic evidence
| note20-at=-0.050 | note20=Modern humans
| note21=
P
l
e
i
s
t
o
c
e
n
e
| note21-at=-0.00 | note21-nudge-left=13 | note21-remove-arrow=yes
| note22=
P
l
i
o
c
e
n
e
| note22-at=-3.50 | note22-nudge-left=13 | note22-remove-arrow=yes
| note23=
M
i
o
c
e
n
e
| note23-at=-6.75 | note23-nudge-left=13 | note23-remove-arrow=yes
| note24=
H
o
m
i
n
i
d
s
| note24-at=-4.20 | note24-nudge-left=0 | note24-nudge-right=-4.20 | note24-remove-arrow=yes
| caption=
}}
Analyses of the modern human genomes indicate past interbreeding with at least two groups of archaic humans, Neanderthals[73] and Denisovans,[41][99] and that such interbreeding events occurred on multiple occasions. Comparisons of the Denisovan, Neanderthal, and modern human genomes have revealed evidence of a complex web of interbreeding among these lineages.[73]
Archaic humans
As much as 17% of the Denisovan genome from Denisova Cave represents DNA from the local Neanderthal population.[73] Denisova 11 was an F1 (first generation) Denisovan/Neanderthal hybrid; the fact that such an individual was found may indicate interbreeding was a common occurrence here.[13] The Denisovan genome shares more derived alleles with the Altai Neanderthal genome from Siberia than with the Vindija Cave Neanderthal genome from Croatia or the Mezmaiskaya cave Neanderthal genome from the Caucasus, suggesting that the gene flow came from a population that was more closely related to the local Altai Neanderthals.[100] However, Denny's Denisovan father had the typical Altai Neanderthal introgression, while her Neanderthal mother represented a population more closely related to Vindija Neanderthals.[101] Denisova 25, dated to 200 ka, is estimated to have inherited 5% of his genome from a previously unknown population of Neanderthals, and came from a different population of Denisovans than the younger samples.[48]
The Denisovan haplotype for the MUC19 gene was also found in the late Neanderthals from Chagyrskaya Cave[102] in the Altai in Siberia and Vindija in Croatia in Europe, which provides proof that late Neanderthals from both Europe and Asia had Denisovan admixture.[103]
About 4% of the Denisovan genome derives from an unidentified archaic hominin,[73] indicating this species diverged from Neanderthals and humans over a million years ago. The only identified Homo species of Late Pleistocene Asia are H. erectus and H. heidelbergensis,[100][104] though in 2021 and 2024, specimens allocated to the latter species were reclassified as H. longi (Denisovan) and H. juluensis.[56][88]
Before splitting from Neanderthals, their ancestors ("Neandersovans") migrating into Europe apparently interbred with an unidentified "superarchaic" human species who were already present there; these superarchaics were the descendants of a very early migration out of Africa around 1.9 mya.[105]
Modern humans

A 2011 study found that Denisovan DNA is present at a comparatively high level in Papuans, Aboriginal Australians, Near Oceanians, Polynesians, Fijians, Eastern Indonesians, and Aeta (from the Philippines); but not in East Asians, western Indonesians, Jahai people (from Malaysia), or Onge (from the Andaman Islands). This may suggest that Denisovan introgression occurred within the Pacific region rather than on the Asian mainland, and that ancestors of the latter groups were not present in Southeast Asia at the time.[83][107] In the Melanesian genome, about 4–6%[41] or 1.9–3.4% derives from Denisovan introgression.[108] Prior to 2021, New Guineans and Indigenous Australians were reported to have the most introgressed DNA,[31] but Australians have less than New Guineans.[109] A 2021 study discovered 30–40% more Denisovan ancestry in Aeta people in the Philippines than in Papuans, estimated as about 5% of the genome. The Aeta Magbukon in Luzon have the highest known proportion of Denisovan ancestry of any population in the world.[86] In Papuans, less Denisovan ancestry is seen in the X chromosome than autosomes, and some autosomes (such as chromosome 11) also have less Denisovan ancestry, which could indicate hybrid incompatibility. The former observation could also be explained by less female Denisovan introgression into modern humans, or more female modern human immigrants who diluted Denisovan X chromosome ancestry.[7]
In contrast, 0.2% derives from Denisovan ancestry in mainland Asians and Native Americans.[10] A 2018 study found that South Asians were found to have levels of Denisovan admixture similar to that seen in East Asians.[11] Another study found that the highest Denisovan ancestry is inferred in Oceanians (~2.0%), while Americans, East Asians, and South Asians have similar amounts (~0.1%).[12] The discovery of the 40,000-year-old Chinese modern human Tianyuan Man lacking Denisovan DNA significantly different from the levels in modern-day East Asians discounts the hypothesis that immigrating modern humans simply diluted Denisovan ancestry whereas Melanesians lived in reproductive isolation.[110][31] A 2018 study of Han Chinese, Japanese, and Dai genomes showed that modern East Asians have DNA from two different Denisovan populations: one similar to the Denisovan DNA found in Papuan genomes, and a second that is closer to the Denisovan genome from Denisova Cave. This could indicate two separate introgression events involving two different Denisovan populations. In South Asian genomes, DNA only came from the same single Denisovan introgression seen in Papuans.[11] A 2019 study found a third wave of Denisovans which introgressed into East Asians. Introgression, also, may not have immediately occurred when modern humans immigrated into the region.[84]
The timing of introgression into Oceanian populations likely occurred after Eurasians and Oceanians split roughly 50,000 years ago,[111] and before Papuan and Aboriginal Australians split from each other roughly 37,000 years ago. Given the present day distribution of Denisovan DNA, this may have taken place in Wallacea, though the discovery of a 7,200 year old Toalean girl (closely related to Papuans and Aboriginal Australians) from Sulawesi carrying Denisovan DNA makes Sundaland another potential candidate. Other early Sunda hunter gatherers so far sequenced carry very little Denisovan DNA, which either means the introgression event did not take place in Sundaland, or Denisovan ancestry was diluted with gene flow from the mainland Asian Hòabìnhian culture and subsequent Neolithic cultures.[112]
A haplotype of EPAS1 in modern Tibetans, which allows them to live at high elevations in a low-oxygen environment, likely came from Denisovans.[92][43] Genes related to phospholipid transporters (which are involved in fat metabolism) and to trace amine-associated receptors (involved in smelling) are more active in people with more Denisovan ancestry.[113] Denisovan genes may have conferred a degree of immunity against the G614 mutation of SARS-CoV-2.[114] Denisovan introgressions may have influenced the immune system of present-day Papuans and potentially favoured "variants to immune-related phenotypes" and "adaptation to the local environment".[115]
In December 2023, scientists reported that genes inherited by modern humans from Neanderthals and Denisovans may biologically influence the daily routine of modern humans.[116]
In August 2025, a study reported an archaic haplotype of the MUC19 gene of Denisovan origin which occurs at high frequency in most admixed American populations, and which was also was found in ancient genomes from 23 ancient Indigenous American individuals who predate admixture with Europeans and Africans.[103] This haplotype regulates MUC19 production. which is expressed in corneal and conjunctival epithelia, and the lacrimal gland[117] and shows strong signs of positive selection in modern humans.[118] This haplotype with Denisovan-specific variants is contained in a 72-kb region of the MUC19 gene, but that region is embedded in a larger 742-kb region that contains Neanderthal-specific variants. The authors also found that the Denisovan haplotype was also found in the Chagyrskaya[102] and Vindija late Neanderthals, so the authors conclude that modern humans inherited this haplotype from Neanderthals, who likely inherited it from Denisovans through Denisovan admixture with Neanderthals.
A 2025 study of ancient and modern genomes from Eurasia traced the historical route of Denisovan admixture throughout Northern and Western Eurasia. It found that the highest amount of Denisovan admixture was in the 40,000 year old Tianyuan man from northeast China, the first example of the Ancestral East Asian population. The individuals from Sungir in Russia from 34,000 years ago also had Denisovan admixture. Tianyuan man contributed Denisovan DNA segments to the individual from Salkhit in Eastern Siberia, and later to the Ancient North Eurasians such as the Mal'ta boy from 24,000 years ago. The Ancient North Eurasians then became the ancestors of the Eastern hunter-gatherers who passed their Denisovan ancestry to the later Proto-Indo-European Yamnaya culture. Therefore, present day Europeans and Near Easterners also have Denisovan ancestry. However, the study found that the Jōmon people of ancient Japan were had much less Denisovan ancestry than all other East Asians.[119]
See also
- Homo longi – Archaic human from China, 146,000 BP
- Homo naledi – South African archaic human species
- Nesher Ramla Homo – Extinct population of archaic humans
- Solo Man – Extinct subspecies of Homo erectus
- Biology:Timeline of human evolution – Chronological outline of major events in the development of the human species
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Fu, Q.; Bai, F.; Rao, H.; Chen, S.; Ji, Y.; Liu, A.; Bennett, E. A.; Liu, F. et al. (2025). "The proteome of the late Middle Pleistocene Harbin individual". Science. doi:10.1126/science.adu9677. PMID 40531192.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Fu, Q.; Cao, P.; Dai, Q.; Bennett, E. A.; Feng, X.; Yang, M. A.; Ping, W.; Pääbo, S. et al. (2025). "Denisovan mitochondrial DNA from dental calculus of the >146,000-year-old Harbin cranium". Cell 188 (15): 3919–3926.e9. doi:10.1016/j.cell.2025.05.040.
- ↑ Curry, Andrew (18 June 2025). "'Dragon Man' skull belongs to mysterious human relative". Science (American Association for the Advancement of Science (AAAS)). doi:10.1126/science.z8sb68w.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Feng, Xiaobo; Yin, Qiyu; Gao, Feng; Lu, Dan; Fang, Qin; Feng, Yilu; Huang, Xuchu; Tan, Chen et al. (25 September 2025). "The phylogenetic position of the Yunxian cranium elucidates the origin of Homo longi and the Denisovans". Science 389 (6767): 1320–1324. doi:10.1126/science.ado9202. https://www.science.org/doi/10.1126/science.ado9202. Retrieved 25 September 2025.
- ↑ 5.0 5.1 5.2 5.3 5.4 Krause, J.; Fu, Q.; Good, J. M.; Viola, B. et al. (2010). "The complete mitochondrial DNA genome of an unknown hominin from southern Siberia" (in en). Nature 464 (7290): 894–897. doi:10.1038/nature08976. ISSN 1476-4687. PMID 20336068. Bibcode: 2010Natur.464..894K.
- ↑ 6.0 6.1 Bower, Bruce (September 2025). "Putting an ancient face to a name". Science News 207 (9): 11.
- ↑ 7.0 7.1 7.2 Meyer, M.Expression error: Unrecognized word "et". (2012). "A High-Coverage Genome Sequence from an Archaic Denisovan Individual". Science 338 (6104): 222–226. doi:10.1126/science.1224344. PMID 22936568. Bibcode: 2012Sci...338..222M.
- ↑ Gokhman, David; Mishol, Nadav; de Manuel, Marc; de Juan, David; Shuqrun, Jonathan; Meshorer, Eran; Marques-Bonet, Tomas; Rak, Yoel et al. (2019-09-19). "Reconstructing Denisovan Anatomy Using DNA Methylation Maps". Cell 179 (1): 180–192. doi:10.1016/j.cell.2019.08.035. PMID 31539495. https://www.cell.com/cell/fulltext/S0092-8674(19)30954-7.
- ↑ Larena, M.; McKenna, J.; Sanchez-Quinto, F.; Bernhardsson, C.; Ebeo, C.; Reyes, R.; Casel, O.; Huang, J.-Y. et al. (11 October 2021). "Philippine Ayta possess the highest level of Denisovan ancestry in the world". Current Biology 31 (19): 4219–4230.e10. doi:10.1016/j.cub.2021.07.022. PMID 34388371. Bibcode: 2021CBio...31E4219L.
- ↑ 10.0 10.1 Prüfer, K.; Racimo, F.; Patterson, N.; Jay, F. et al. (2013). "The complete genome sequence of a Neanderthal from the Altai Mountains". Nature 505 (7481): 43–49. doi:10.1038/nature12886. PMID 24352235. Bibcode: 2014Natur.505...43P.
- ↑ 11.0 11.1 11.2 Browning, S. R.; Browning, B. L.; Zhou, Yi.; Tucci, S. et al. (2018). "Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture". Cell 173 (1): 53–61.e9. doi:10.1016/j.cell.2018.02.031. ISSN 0092-8674. PMID 29551270.
- ↑ 12.0 12.1 Elise Kerdoncuff; Laurits Skov; Nick Patterson; Joyita Banerjee; Pranali Khobragade; Sankha S. Chakrabarti; Avinash Chakrawarty; Prasun Chatterjee et al. (26 June 2025). "50,000 years of evolutionary history of India: Impact on health and disease variation". The Innovation 188 (13): 3389–3404.e6. doi:10.1016/j.cell.2025.04.027. PMID 40578318. https://www.cell.com/cell/fulltext/S0092-8674(25)00462-3.
- ↑ 13.0 13.1 Warren, M. (2018). "Mum's a Neanderthal, Dad's a Denisovan: First discovery of an ancient-human hybrid – Genetic analysis uncovers a direct descendant of two different groups of early humans.". Nature 560 (7719): 417–418. doi:10.1038/d41586-018-06004-0. PMID 30135540. Bibcode: 2018Natur.560..417W.
- ↑ Vogel, Gretchen (22 August 2018). "This ancient bone belonged to a child of two extinct human species". Science. doi:10.1126/science.aav1858. https://www.science.org/content/article/ancient-bone-belonged-child-two-extinct-human-species. Retrieved 22 August 2018.
- ↑ Rogers, A. R.; Harris, N. S.; Achenbach, A. A. (21 February 2020). "Neanderthal-Denisovan ancestors interbred with a distantly related hominin". Science Advances 6 (8). doi:10.1126/sciadv.aay5483. PMID 32128408. Bibcode: 2020SciA....6.5483R.
- ↑ Welker, Frido; Ramos-Madrigal, Jazmín; Gutenbrunner, Petra; Mackie, Meaghan; Tiwary, Shivani; Rakownikow Jersie-Christensen, Rosa; Chiva, Cristina; Dickinson, Marc R. et al. (2020). "The dental proteome of Homo antecessor". Nature 580 (7802): 235–238. doi:10.1038/s41586-020-2153-8. ISSN 1476-4687. PMID 32269345. Bibcode: 2020Natur.580..235W.
- ↑ Madupe, Palesa P.; Koenig, Claire; Patramanis, Patramanis; Rüther, Patrick L.; Hlazo, Nomawethu; Mackie, Meaghan; Tawane, Mirriam; Krueger, Johanna et al. (29 May 2025). "Enamel proteins reveal biological sex and genetic variability in southern African Paranthropus". Science 388 (6750): 969–973. doi:10.1126/science.adt9539. PMID 40440366. PMC 7617798. https://www.science.org/doi/10.1126/science.adt9539.
- ↑ Derevianko, A.P. (September 2011). "The Origin of Anatomically Modern Humans and their Behavior in Africa and Eurasia". Archaeology, Ethnology and Anthropology of Eurasia 39 (3): 2–31. doi:10.1016/j.aeae.2011.09.001.
- ↑ Picq, P. (24 November 2011). L'homme est-il un grand singe politique?. Paris: Odile Jacob. p. 24. ISBN 978-2-7381-2698-6.
- ↑ Gabriel, C.C.; Mihaela, C. (December 2011). "Considerations on human evolution and on species origin centers". Oltenia, Studii Si Comunicari, Stiintele Naturii 27 (2): 210–217. http://olteniastudiisicomunicaristiintelenaturii.ro/v27_2.html.
- ↑ Gunbin, K.V.; Afonnikov, D.A.; Kolchanov, N.A.; Derevianko, A.P. (September 2012). "The Importance of Changes to microRNA in the Evolution of Homo neanderthalensis and Homo denisova". Archaeology, Ethnology and Anthropology of Eurasia 40 (3): 22–30. doi:10.1016/j.aeae.2012.11.004.
- ↑ Reed, D. (2025). "Nomenclature and Taxonomy of Chibanian Hominins". PaleoAnthropology 2: 1–14. https://paleoanthropology.org/ojs/index.php/paleo/libraryFiles/downloadPublic/33.
- ↑ Manoa, University of Hawaii at. "Homo juluensis: Possible new ancient human species uncovered by researchers" (in en). https://phys.org/news/2024-11-homo-juluensis-ancient-human-species.html.
- ↑ Bae, Christopher J.; Wu, Xiujie (2024-11-02). "Making sense of eastern Asian Late Quaternary hominin variability" (in en). Nature Communications 15 (1): 9479. doi:10.1038/s41467-024-53918-7. ISSN 2041-1723. PMID 39488555. Bibcode: 2024NatCo..15.9479B.
- ↑ 25.0 25.1 Callaway, Ewen (2010). "Fossil genome reveals ancestral link". Nature 468 (7327): 1012. doi:10.1038/4681012a. PMID 21179140. Bibcode: 2010Natur.468.1012C.
- ↑ Ao, H.; Liu, C.-R.; Roberts, A. P. (2017). "An updated age for the Xujiayao hominin from the Nihewan Basin, North China: Implications for Middle Pleistocene human evolution in East Asia". Journal of Human Evolution 106: 54–65. doi:10.1016/j.jhevol.2017.01.014. PMID 28434540. Bibcode: 2017AGUFMPP13B1080A.
- ↑ Li, Zhan-Yang; Wu, Xiu-Jie; Zhou, Li-Ping; Liu, Wu; Gao, Xing; Nian, Xiao-Mei; Trinkaus, Erik (3 Mar 2017). "Late Pleistocene archaic human crania from Xuchang, China". Science 355 (6328): 969–972. doi:10.1126/science.aal2482. PMID 28254945. Bibcode: 2017Sci...355..969L. https://www.science.org/doi/10.1126/science.aal2482.
- ↑ Lewis, Dyani (2023-09-18). "A new human species? Mystery surrounds 300,000-year-old fossil" (in en). Nature. doi:10.1038/d41586-023-02924-8. PMID 37723275. https://www.nature.com/articles/d41586-023-02924-8.
- ↑ Li, Tianyuan; Etler, Dennis A. (4 June 1992). "New Middle Pleistocene hominid crania from Yunxian in China" (in en). Nature 357 (6377): 404–407. doi:10.1038/357404a0. ISSN 1476-4687. PMID 1594044. Bibcode: 1992Natur.357..404T. https://www.nature.com/articles/357404a0. Retrieved 6 May 2024.
- ↑ Woo, Ju-Kang; Peng, Ru-Ce (December 1959). "Fossil Human Skull of Early Paleoanthropic Stage Found at Mapa, Shaoquan, Kwangtung Province". Vertebrata PalAsiatica 3 (4): 176–182. http://www.ivpp.cas.cn/cbw/gjzdwxb/xbwzml/201106/P020110622319912556127.pdf.
- ↑ 31.0 31.1 31.2 31.3 31.4 Cooper, A.; Stringer, C. B. (2013). "Did the Denisovans Cross Wallace's Line?". Science 342 (6156): 321–23. doi:10.1126/science.1244869. PMID 24136958. Bibcode: 2013Sci...342..321C.
- ↑ 32.0 32.1 32.2 32.3 32.4 32.5 Ji, Qiang; Wu, Wensheng; Ji, Yannan; Li, Qiang; Ni, Xijun (2021-06-25). "Late Middle Pleistocene Harbin cranium represents a new Homo species" (in en). The Innovation 2 (3). doi:10.1016/j.xinn.2021.100132. ISSN 2666-6758. PMID 34557772. Bibcode: 2021Innov...200132J.
- ↑ Bae, C. J. (2024). "The "Muddle in the Middle" (~400 ka–~100 ka)". The Paleoanthropology of Eastern Asia. University of Hawaiʻi Press. pp. 95–131. doi:10.1515/9780824898106-007. ISBN 978-0-8248-9810-6.
- ↑ Laiu, Darryl (6 January 2025). "Homo juluensis: Possible 'new ancient human' identified". BBC. https://www.bbc.com/reel/video/p0kftjyg/homo-juluensis-possible-new-ancient-human-identified.
- ↑ Ovodov, N. D.; Crockford, S. J.; Kuzmin, Y. V.; Higham, T. F. et al. (2011). "A 33,000-Year-Old Incipient Dog from the Altai Mountains of Siberia: Evidence of the Earliest Domestication Disrupted by the Last Glacial Maximum". PLOS ONE 6 (7). doi:10.1371/journal.pone.0022821. PMID 21829526. Bibcode: 2011PLoSO...622821O.
- ↑ Reich, D. (2018). Who We Are and How We Got Here. Oxford University Press. p. 53. ISBN 978-0-19-882125-0.
- ↑ 37.0 37.1 37.2 37.3 37.4 37.5 37.6 Douka, K. (2019). "Age estimates for hominin fossils and the onset of the Upper Palaeolithic at Denisova Cave". Nature 565 (7741): 640–644. doi:10.1038/s41586-018-0870-z. PMID 30700871. Bibcode: 2019Natur.565..640D. https://ro.uow.edu.au/cgi/viewcontent.cgi?article=1559&context=smhpapers1. Retrieved 7 December 2019.
- ↑ 38.0 38.1 38.2 38.3 38.4 38.5 Jacobs, Zenobia; Li, Bo; Shunkov, Michael V.; Kozlikin, Maxim B. et al. (January 2019). "Timing of archaic hominin occupation of Denisova Cave in southern Siberia". Nature 565 (7741): 594–599. doi:10.1038/s41586-018-0843-2. ISSN 1476-4687. PMID 30700870. Bibcode: 2019Natur.565..594J. https://ro.uow.edu.au/cgi/viewcontent.cgi?article=1558&context=smhpapers1. Retrieved 29 May 2020.
- ↑ 39.0 39.1 39.2 39.3 39.4 39.5 Brown, S; Massilani, D; Kozlikin, MB; Shunkov, MV; Derevianko, AP; Stoessel, A; Jope-Street, B; Meyer, M et al. (January 2022). "The earliest Denisovans and their cultural adaptation.". Nature Ecology & Evolution 6 (1): 28–35. doi:10.1038/s41559-021-01581-2. PMID 34824388.
- ↑ 40.0 40.1 40.2 Cite error: Invalid
<ref>tag; no text was provided for refs namedSlon Viola Renaud Gansauge 2017 - ↑ 41.0 41.1 41.2 41.3 41.4 41.5 41.6 41.7 Reich, D.Expression error: Unrecognized word "et". (2010). "Genetic history of an archaic hominin group from Denisova Cave in Siberia". Nature 468 (7327): 1053–60. doi:10.1038/nature09710. PMID 21179161. PMC 4306417. Bibcode: 2010Natur.468.1053R. http://repositori.upf.edu/bitstream/10230/25596/1/Marques_nat_gen.pdf. Retrieved 29 July 2018.
- ↑ Gibbons, Anne (2019). "First fossil jaw of Denisovans finally puts a face on elusive human relatives". Science. doi:10.1126/science.aax8845.
- ↑ 43.0 43.1 43.2 43.3 43.4 43.5 43.6 Chen, F.Expression error: Unrecognized word "et". (2019). "A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau". Nature 569 (7756): 409–412. doi:10.1038/s41586-019-1139-x. PMID 31043746. Bibcode: 2019Natur.569..409C. https://kar.kent.ac.uk/74280/1/Xiahe_Main.pdf. Retrieved 7 December 2019.
- ↑ Shang, D. (2020). "Denisovan DNA in Late Pleistocene sediments from Baishiya Karst Cave on the Tibetan Plateau". Science 370 (6516): 584–587. doi:10.1126/science.abb6320. PMID 33122381.
- ↑ 45.0 45.1 Xia, Huan; Zhang, Dongju; Wang, Jian; Fagernäs, Zandra; Li, Ting; Li, Yuanxin; Yao, Juanting; Lin, Dongpeng et al. (3 July 2024). "Middle and Late Pleistocene Denisovan subsistence at Baishiya Karst Cave" (in en). Nature 632 (8023): 108–113. doi:10.1038/s41586-024-07612-9. ISSN 1476-4687. PMID 38961285. Bibcode: 2024Natur.632..108X.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedXiahe2 - ↑ 47.0 47.1 47.2 47.3 47.4 Demeter, F.Expression error: Unrecognized word "et". (2022). "A Middle Pleistocene Denisovan molar from the Annamite Chain of northern Laos". Nature Communications 13 (2557): 2557. doi:10.1038/s41467-022-29923-z. PMID 35581187. Bibcode: 2022NatCo..13.2557D.
- ↑ 48.0 48.1 48.2 48.3 Gibbons, Ann (2024-07-11). "The most ancient human genome yet has been sequenced—and it's a Denisovan's". Science. doi:10.1126/science.zi9n4zp. https://www.science.org/content/article/most-ancient-human-genome-yet-has-been-sequenced-and-it-s-denisovan. Retrieved 13 July 2024.
- ↑ 49.0 49.1 Sawyer, S.; Renaud, G.; Viola, B.; Hublin, J.-J. et al. (2015). "Nuclear and mitochondrial DNA sequences from two Denisovan individuals". Proceedings of the National Academy of Sciences 112 (51): 15696–700. doi:10.1073/pnas.1519905112. PMID 26630009. Bibcode: 2015PNAS..11215696S.
- ↑ 50.0 50.1 50.2 Bennett, E. A.Expression error: Unrecognized word "et". (2019). "Morphology of the Denisovan phalanx closer to modern humans than to Neanderthals". Science Advances 5 (9). doi:10.1126/sciadv.aaw3950. PMID 31517046. Bibcode: 2019SciA....5.3950B.
- ↑ 51.0 51.1 51.2 51.3 Tsutaya, Takumi; Sawafuji, Rikai; Taurozzi, Alberto J.; Fagernäs, Zandra; Patramanis, Ioannis; Troché, Gaudry; Mackie, Meaghan; Gakuhari, Takashi et al. (2025-04-11). "A male Denisovan mandible from Pleistocene Taiwan" (in en). Science 388 (6743): 176–180. doi:10.1126/science.ads3888. ISSN 0036-8075. PMID 40208980. Bibcode: 2025Sci...388..176T. https://www.science.org/doi/10.1126/science.ads3888.
- ↑ Chang, Chun-Hsiang; Kaifu, Yousuke; Takai, Masanaru; Kono, Reiko T.; Grün, Rainer; Matsu'ura, Shuji; Kinsley, Les; Lin, Liang-Kong (2015). "The first archaic Homo from Taiwan". Nature Communications 6. doi:10.1038/ncomms7037. PMID 25625212. Bibcode: 2015NatCo...6.6037C.
- ↑ Viola, B. T.; Gunz, P.; Neubauer, S. (2019). "A parietal fragment from Denisova cave". 88th Annual Meeting of the American Association of Physical Anthropologists. https://meeting.physanth.org/program/2019/session09/viola-2019-a-parietal-fragment-from-denisova-cave.html. Retrieved 18 January 2020.
- ↑ Jacobs, Z.; Zavala, E. I.; Li, B.; O'Gorman, K.; Shunkov, M. V.; Kozlikin, M. B.; Derevianko, A. P.; Uliyanov, V. A. et al. (2025). "Pleistocene chronology and history of hominins and fauna at Denisova Cave". Nature Communications 16 (1): 4738. doi:10.1038/s41467-025-60140-6. PMID 40399313. Bibcode: 2025NatCo..16.4738J.
- ↑ Lewis, Dyani (2025-06-18). "First ever skull from 'Denisovan' reveals what ancient people looked like" (in en). Nature 642 (8069): 848–849. doi:10.1038/d41586-025-01899-y. ISSN 1476-4687. PMID 40537588. Bibcode: 2025Natur.642..848L. https://www.nature.com/articles/d41586-025-01899-y. "Qiang Ji, a palaeontologist at Hebei GEO University in Shijiazhuang, China, obtained the specimen from an unnamed man in 2018. The man — who Ji suspects discovered the artefact himself but failed to report it to authorities — claimed that his grandfather unearthed the fossil in 1933 during bridge-construction work over Long Jiang (which means dragon river), and buried it in an abandoned well, where it remained until a deathbed confession.".
- ↑ 56.0 56.1 56.2 56.3 56.4 56.5 56.6 Ni, X.Expression error: Unrecognized word "et". (2021). "Massive cranium from Harbin in northeastern China establishes a new Middle Pleistocene human lineage". The Innovation 2 (3). doi:10.1016/j.xinn.2021.100130. ISSN 2666-6758. PMID 34557770. Bibcode: 2021Innov...200130N.
- ↑ Wu, X. Z. (1981). "A well-preserved cranium of an archaic type of early Homo sapiens from Dali, China". Scientia Sinica 24 (4): 530–41. PMID 6789450.
- ↑ Xuefeng Sun et al. , "TT-OSL and post-IR IRSL dating of the Dali Man site in central China", Quaternary International 434, Part A, 1 April 2017, 99-106, doi:10.1016/j.quaint.2015.05.027 "correlating the pIRIR290 ages between 267.7 ± 13.9 ka and 258.3 ± 14.2 ka and new pollen analysis, we proposed a new viewpoint that the Dali Man was likely to live during a transitional period from glacial to interglacial climate in the S2/L3 (MIS 7/8) stage."
- ↑ Guoqin, Qi (2023-04-04). "The pleistocene human environment of North China" (in en-US). Acta Anthropologica Sinica 9 (4): 340. ISSN 1000-3193. https://www.sciengine.com/A-A-S/doi/10.0000/1000-3193-9/04/340.
- ↑ Tattersall, Ian; Schwartz, Jeffrey H. (2009-05-30). "Evolution of the Genus Homo" (in en). Annual Review of Earth and Planetary Sciences 37 (1): 67–92. doi:10.1146/annurev.earth.031208.100202. ISSN 0084-6597. Bibcode: 2009AREPS..37...67T. https://www.annualreviews.org/content/journals/10.1146/annurev.earth.031208.100202.
- ↑ Bower, Bruce (2006-02-22). "Big Woman with a Distant Past: Stone Age gal embodies humanity's cold shifts" (in en-US). https://www.sciencenews.org/article/big-woman-distant-past-stone-age-gal-embodies-humanitys-cold-shifts.
- ↑ Liu, Wu; Wu, Xiujie (23 Mar 2011). "The Hominid Fossils from China Contemporaneous with the Neanderthals and Some Related Studies". in Condemi, Silvana; Weniger, Gerd-Christian. Continuity and Discontinuity in the Peopling of Europe: One Hundred Fifty Years of Neanderthal Study. Springer Science & Business Media. ISBN 978-94-007-0491-6. https://books.google.com/books?id=i75qyoRhyg8C&pg=PA77.
- ↑ Xing, Song; Martinón-Torres, María; Bermúdez de Castro, Jose María; Wu, Xiujie; Liu, Wu (2015). "Hominin teeth from the early Late Pleistocene site of Xujiayao, Northern China". American Journal of Physical Anthropology 156 (2): 224–240. doi:10.1002/ajpa.22641. PMID 25329008.
- ↑ Xing, Song; Martinón-Torres, María; María Bermúdez de Castro, Jose; Wu, Xiujie; Liu, Wu (2014). "Hominin teeth from the early Late Pleistocene site of Xujiayao, Northern China". American Journal of Physical Anthropology 156 (2): 224-240. doi:10.1002/ajpa.22641. http://onlinelibrary.wiley.com/doi/10.1002/ajpa.22641.
- ↑ Wu, Xiu-Jie; Pei, Shu-Wen; Cai, Yan-Jun; Tong, Hao-Wen; Li, Qiang; Dong, Zhe; Sheng, Jin-Chao; Jin, Ze-Tian et al. (2019-05-14). "Archaic human remains from Hualongdong, China, and Middle Pleistocene human continuity and variation". Proceedings of the National Academy of Sciences of the United States of America 116 (20): 9820–9824. doi:10.1073/pnas.1902396116. ISSN 1091-6490. PMID 31036653. Bibcode: 2019PNAS..116.9820W.
- ↑ 66.0 66.1 Wu, Xiujie; Pei, Shuwen; Cai, Yanjun; Tong, Haowen; Zhang, Ziliang; Yan, Yi; Xing, Song; Martinón-Torres, María et al. (2023). "Morphological and morphometric analyses of a late Middle Pleistocene hominin mandible from Hualongdong, China". Journal of Human Evolution 182. doi:10.1016/j.jhevol.2023.103411. PMID 37531709. Bibcode: 2023JHumE.18203411W. https://www.sciencedirect.com/science/article/pii/S0047248423000908.
- ↑ 67.0 67.1 67.2 Woo, Ju-Kang; Peng, Ru-Ce (1959). "Guangdong shaoguan maba faxian de zaoqi gurenleixing renlei huashi". Vertebrata PalAsiatica 3: 176–182. http://www.ivpp.cas.cn/cbw/gjzdwxb/xbwzxz/201106/P020110622319912556127.pdf.
- ↑ Wu, Xiu-jie; Bruner, Emiliano (2016-03-12). "The endocranial anatomy of maba 1". American Journal of Physical Anthropology 160 (4): 633–643. doi:10.1002/ajpa.22974. ISSN 0002-9483. PMID 26972814. Bibcode: 2016AJPA..160..633W. http://dx.doi.org/10.1002/ajpa.22974.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedPeng1959 - ↑ Lao, O.; Bertranpetit, J.; Mondal, M. (2019). "Approximate Bayesian computation with deep learning supports a third archaic introgression in Asia and Oceania" (in en). Nature Communications 10 (1): 246. doi:10.1038/s41467-018-08089-7. ISSN 2041-1723. PMID 30651539. Bibcode: 2019NatCo..10..246M.
- ↑ Rogers, A. R.; Bohlender, R. J.; Huff, C. D. (2017). "Early history of Neanderthals and Denisovans". Proceedings of the National Academy of Sciences 114 (37): 9859–9863. doi:10.1073/pnas.1706426114. PMID 28784789. Bibcode: 2017PNAS..114.9859R.
- ↑ Ho, K. K. (2016). "Hominin interbreeding and the evolution of human variation". Journal of Biological Research-Thessaloniki 23. doi:10.1186/s40709-016-0054-7. PMID 27429943.
- ↑ 73.0 73.1 73.2 73.3 73.4 Pennisi, E. (2013). "More Genomes from Denisova Cave Show Mixing of Early Human Groups". Science 340 (6134): 799. doi:10.1126/science.340.6134.799. PMID 23687020. Bibcode: 2013Sci...340..799P.
- ↑ Pääbo, S.; Kelso, J.; Reich, D.; Slatkin, M. et al. (2014). "The complete genome sequence of a Neanderthal from the Altai Mountains" (in en). Nature 505 (7481): 43–49. doi:10.1038/nature12886. ISSN 1476-4687. PMID 24352235. Bibcode: 2014Natur.505...43P.
- ↑ Kuhlwilm, M.; Gronau, I.; Hubisz, M. J.; de Filippo, C. et al. (2016). "Ancient gene flow from early modern humans into Eastern Neanderthals". Nature 530 (7591): 429–433. doi:10.1038/nature16544. ISSN 1476-4687. PMID 26886800. Bibcode: 2016Natur.530..429K.
- ↑ Posth, C.; Wißing, C.; Kitagawa, K.; Pagani, L. et al. (2017). "Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals". Nature Communications 8. doi:10.1038/ncomms16046. ISSN 2041-1723. PMID 28675384. Bibcode: 2017NatCo...816046P.
- ↑ Bertranpetit, J.; Majumder, P. P.; Li, Q.; Laayouni, H. et al. (2016). "Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation". Nature Genetics 48 (9): 1066–1070. doi:10.1038/ng.3621. ISSN 1546-1718. PMID 27455350.
- ↑ Callaway, E. (2013). "Hominin DNA baffles experts". Nature 504 (7478): 16–17. doi:10.1038/504016a. PMID 24305130. Bibcode: 2013Natur.504...16C.
- ↑ Tattersall, I. (2015). The Strange Case of the Rickety Cossack and other Cautionary Tales from Human Evolution. Palgrave Macmillan. p. 200. ISBN 978-1-137-27889-0.
- ↑ Meyer, M.Expression error: Unrecognized word "et". (2016). "Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins". Nature 531 (7595): 504–507. doi:10.1038/nature17405. PMID 26976447. Bibcode: 2016Natur.531..504M.
- ↑ Petr, Martin; Hajdinjak, Mateja; Fu, Qiaomei; Essel, Elena; Rougier, Hélène; Crevecoeur, Isabelle; Semal, Patrick; Golovanova, Liubov V. et al. (25 Sep 2020). "The evolutionary history of Neanderthal and Denisovan Y chromosomes". Science 369 (6511): 1653–1656. doi:10.1126/science.abb6460. PMID 32973032. Bibcode: 2020Sci...369.1653P. https://www.science.org/doi/10.1126/science.abb6460. Retrieved 3 August 2025.
 This article incorporates text from this source, which is available under the CC BY 4.0 license.
This article incorporates text from this source, which is available under the CC BY 4.0 license.
- ↑ Callaway, E. (2011). "First Aboriginal genome sequenced". Nature News. doi:10.1038/news.2011.551.
- ↑ 83.0 83.1 83.2 Reich, David; Patterson, Nick; Kircher, Martin; Delfin, Frederick et al. (2011). "Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania". The American Journal of Human Genetics 89 (4): 516–28. doi:10.1016/j.ajhg.2011.09.005. PMID 21944045.
- ↑ 84.0 84.1 84.2 84.3 Jacobs, G. S.; Hudjashov, G.; Saag, L.; Kusuma, P. et al. (2019). "Multiple Deeply Divergent Denisovan Ancestries in Papuans". Cell 177 (4): 1010–1021.e32. doi:10.1016/j.cell.2019.02.035. ISSN 0092-8674. PMID 30981557.
- ↑ "Denisovan DNA may help modern humans adapt to different environments". https://www.newscientist.com/article/2438941-denisovan-dna-may-help-modern-humans-adapt-to-different-environments/.
- ↑ 86.0 86.1 Larena, Maximilian; McKenna, James; Sanchez-Quinto, Federico; Bernhardsson, Carolina et al. (August 2021). "Philippine Ayta possess the highest level of Denisovan ancestry in the world". Current Biology 31 (19): 4219–4230.e10. doi:10.1016/j.cub.2021.07.022. PMID 34388371. Bibcode: 2021CBio...31E4219L.
- ↑ Warren, M. (2019). "Biggest Denisovan fossil yet spills ancient human's secrets". Nature News 569 (7754): 16–17. doi:10.1038/d41586-019-01395-0. PMID 31043736. Bibcode: 2019Natur.569...16W.
- ↑ 88.0 88.1 88.2 88.3 Bae, C. J.; Wu, X. (2024). "Making sense of eastern Asian Late Quaternary hominin variability". Nature Communications 15 (9479): 9479. doi:10.1038/s41467-024-53918-7. PMID 39488555. Bibcode: 2024NatCo..15.9479B.
- ↑ Bermudez de Castro, J.M.; Arsuaga, J.L.; Carbonell, E.; Rosas, A.; Martinez, I.; Mosquera, M. (1997). "A hominid from the Lower Pleistocene of Atapuerca, Spain: possible ancestor to Neandertals and modern humans". Science 276 (5317): 1392–1395. doi:10.1126/science.276.5317.1392. PMID 9162001.
- ↑ Wu, Rukang (2023-04-04). "The reconstruction of the fossil human skull from Jinniushan, Yinkou, Liaoning Province and its maintures" (in en-US). Acta Anthropologica Sinica 7 (2): 97–101. ISSN 1000-3193. https://www.anthropol.ac.cn/EN/Y1988/V7/I02/97.
- ↑ 91.0 91.1 Rosenberg, Karen R.; Zuné, Lü; Ruff, Christopher B. (2006-03-07). "Body size, body proportions, and encephalization in a Middle Pleistocene archaic human from northern China". Proceedings of the National Academy of Sciences 103 (10): 3552–3556. doi:10.1073/pnas.0508681103. PMID 16505378. Bibcode: 2006PNAS..103.3552R.
- ↑ 92.0 92.1 Huerta-Sánchez, E.Expression error: Unrecognized word "et". (2014). "Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA". Nature 512 (7513): 194–97. doi:10.1038/nature13408. PMID 25043035. Bibcode: 2014Natur.512..194H.
- ↑ Racimo, Fernando; Gokhman, David; Fumagalli, Matteo; Ko, Amy et al. (2017). "Archaic Adaptive Introgression in TBX15/WARS2". Molecular Biology and Evolution 34 (3): 509–524. doi:10.1093/molbev/msw283. PMID 28007980.
- ↑ Dennell, R. (2019). "Dating of hominin discoveries at Denisova". Nature News 565 (7741): 571–572. doi:10.1038/d41586-019-00264-0. PMID 30700881. Bibcode: 2019Natur.565..571D.
- ↑ 95.0 95.1 Zhang, D. D.Expression error: Unrecognized word "et". (2021). "Earliest parietal art: Hominin hand and foot traces from the middle Pleistocene of Tibet". Science Bulletin 66 (24): 2506–2515. doi:10.1016/j.scib.2021.09.001. ISSN 2095-9273. PMID 36654210. Bibcode: 2021SciBu..66.2506Z.
- ↑ Taub, Benjamin (2025-07-04). "300,000-Year-Old Wooden Tools "Made By Denisovans" Discovered In China". IFL Science. https://www.iflscience.com/300000-year-old-wooden-tools-made-by-denisovans-discovered-in-china-79857.
- ↑ Zhang, Dongju (30 Oct 2020). "Denisovan DNA in Late Pleistocene sediments from Baishiya Karst Cave on the Tibetan Plateau". Science 370 (6516): 584–587. doi:10.1126/science.abb6320. PMID 33122381. https://www.science.org/doi/10.1126/science.abb6320.
- ↑ Liu, Jian-Hui; Ge, Jun-Yi; Huang, Yong-Jiang et al. (3 Jul 2025). "300,000-year-old wooden tools from Gantangqing, southwest China". Science 389 (6755): 78–83. doi:10.1126/science.adr8540. PMID 40608932. Bibcode: 2025Sci...389...78L. https://www.science.org/doi/10.1126/science.adr8540.
- ↑ "A draft sequence of the Neandertal genome". Science 328 (5979): 710–22. 2010. doi:10.1126/science.1188021. PMID 20448178. PMC 5100745. Bibcode: 2010Sci...328..710G. http://www.eva.mpg.de/neandertal/press/presskit-neandertal/pdf/Science_Green.pdf. Retrieved 3 May 2013.
- ↑ 100.0 100.1 Prüfer, K.Expression error: Unrecognized word "et". (2013). "The complete genome sequence of a Neanderthal from the Altai Mountains". Nature 505 (7481): 43–49. doi:10.1038/nature12886. PMID 24352235. Bibcode: 2014Natur.505...43P.
- ↑ Warren, Matthew (2018). "Mum's a Neanderthal, Dad's a Denisovan: First discovery of an ancient-human hybrid". Nature 560 (7719): 417–18. doi:10.1038/d41586-018-06004-0. PMID 30135540. Bibcode: 2018Natur.560..417W.
- ↑ 102.0 102.1 Mafessoni, Fabrizio; Grote, Steffi; de Filippo, Cesare; Slon, Viviane; Kolobova, Kseniya A.; Viola, Bence; Markin, Sergey V.; Chintalapati, Manjusha et al. (2020-06-30). "A high-coverage Neandertal genome from Chagyrskaya Cave" (in en). Proceedings of the National Academy of Sciences 117 (26): 15132–15136. doi:10.1073/pnas.2004944117. ISSN 0027-8424. PMID 32546518. PMC 7334501. https://pnas.org/doi/full/10.1073/pnas.2004944117.
- ↑ 103.0 103.1 Villanea, Fernando A.; Peede, David; Kaufman, Eli J.; Añorve-Garibay, Valeria; Chevy, Elizabeth T.; Villa-Islas, Viridiana; Witt, Kelsey E.; Zeloni, Roberta et al. (21 Aug 2025). "The MUC19 gene: An evolutionary history of recurrent introgression and natural selection". Science 389 (6762). doi:10.1126/science.adl0882. PMID 37808839. PMC 10557577. https://www.science.org/doi/10.1126/science.adl0882.
- ↑ Wolf, A. B.; Akey, J. M. (2018). "Outstanding questions in the study of archaic hominin admixture". PLOS Genetics 14 (5). doi:10.1371/journal.pgen.1007349. PMID 29852022.
- ↑ Rogers, A. R.; Harris, N. S.; Achenbach, A. A. (2020). "Neanderthal-Denisovan ancestors interbred with a distantly related hominin". Science Advances 6 (8). doi:10.1126/sciadv.aay5483. PMID 32128408. Bibcode: 2020SciA....6.5483R.
- ↑ Yuan, Kai; Ni, Xumin; Liu, Chang; Pan, Yuwen; Deng, Lian; Zhang, Rui; Gao, Yang; Ge, Xueling et al. (29 October 2021). "Refining models of archaic admixture in Eurasia with ArchaicSeeker 2.0". Nature Communications 12 (1). doi:10.1038/s41467-021-26503-5. PMID 34716342. Bibcode: 2021NatCo..12.6232Y.
- ↑ Yang, Melinda A. (2022-01-06). "A genetic history of migration, diversification, and admixture in Asia" (in en). Human Population Genetics and Genomics 2 (1): 1–32. doi:10.47248/hpgg2202010001. ISSN 2770-5005. https://www.pivotscipub.com/hpgg/2/1/0001. Retrieved 5 December 2023.
- ↑ Vernot, B. (2016). "Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals". Science 352 (6282): 235–239. doi:10.1126/science.aad9416. PMID 26989198. Bibcode: 2016Sci...352..235V.
- ↑ Rasmussen, M.Expression error: Unrecognized word "et". (2011). "An Aboriginal Australian genome reveals separate human dispersals into Asia". Science 334 (6052): 94–98. doi:10.1126/science.1211177. PMID 21940856. Bibcode: 2011Sci...334...94R.
- ↑ Fu, Q.Expression error: Unrecognized word "et". (2013). "DNA analysis of an early modern human from Tianyuan Cave, China". Proceedings of the National Academy of Sciences 110 (6): 2223–2227. doi:10.1073/pnas.1221359110. PMID 23341637. Bibcode: 2013PNAS..110.2223F.
- ↑ Allen, Jim; O'Connell, James F. (29 June 2025). "Recent DNA Studies Question a 65 kya Arrival of Humans in Sahul". Archaeology in Oceania 60 (2): 187–190. doi:10.1002/arco.70002. https://onlinelibrary.wiley.com/doi/full/10.1002/arco.70002?campaign=wolearlyview.
- ↑ Carlhoff, Selina (2021). "Genome of a middle Holocene hunter-gatherer from Wallace". Nature 596 (7873): 543–547. doi:10.1038/s41586-021-03823-6. PMID 34433944. Bibcode: 2021Natur.596..543C.
- ↑ Sankararaman, S.; Mallick, S.; Patterson, N.; Reich, D. (2016). "The combined landscape of Denisovan and Neanderthal ancestry in present-day humans". Current Biology 26 (9): 1241–1247. doi:10.1016/j.cub.2016.03.037. PMID 27032491. Bibcode: 2016CBio...26.1241S.
- ↑ Muscat, Baron Y. (2021). "Could the Denisovan Genes have conferred enhanced Immunity Against the G614 Mutation of SARS-CoV-2?". Human Evolution 36. https://www.researchgate.net/publication/352888303. Retrieved 19 July 2021.
- ↑ Vespasiani, Davide M.; Jacobs, Guy S.; Cook, Laura E.; Brucato, Nicolas; Leavesley, Matthew; Kinipi, Christopher; Ricaut, François-Xavier; Cox, Murray P. et al. (2022-12-08). "Denisovan introgression has shaped the immune system of present-day Papuans". PLOS Genetics 18 (12). doi:10.1371/journal.pgen.1010470. ISSN 1553-7390. PMID 36480515.
- ↑ Zimmer, Carl (14 December 2023). "Morning Person? You Might Have Neanderthal Genes to Thank. - Hundreds of genetic variants carried by Neanderthals and Denisovans are shared by people who like to get up early.". The New York Times. https://www.nytimes.com/2023/12/14/science/neanderthal-sleep-morning-people.html.
- ↑ Martinez-Carrasco, Rafael; Argüeso, Pablo; Fini, M. Elizabeth (2021-07-01). "Membrane-associated mucins of the human ocular surface in health and disease" (in en). The Ocular Surface 21: 313–330. doi:10.1016/j.jtos.2021.03.003. ISSN 1542-0124. PMID 33775913.
- ↑ Hawks, John (2025-08-22). "The gene from Denisovan to Neanderthal to modern mucus". https://www.johnhawks.net/p/the-gene-from-denisovan-to-neanderthal.
- ↑ Yang, Jiaqi; Iasi, Leonardo N. M.; Fu, Qiaomei; Cooke, Niall P.; Kelso, Janet; Peyrégne, Stéphane; Peter, Benjamin M. (2025-10-20). "An early East Asian lineage with unexpectedly low Denisovan ancestry" (in English). Current Biology 35 (20): 4898–4908.e4. doi:10.1016/j.cub.2025.08.051. ISSN 0960-9822. https://www.cell.com/current-biology/abstract/S0960-9822(25)01117-0.
Further reading
- Karlsson, Mattis. From Fossil To Fact: The Denisova Discovery as Science in Action (Thesis). LiU E-press. ISBN 978-91-7929-171-6. Retrieved 18 March 2022.
- Condemi, Silvana and Savatier, François (2025). The Secret World of Denisovans: The Epic Story of the Ancient Cousins to Sapiens and Neanderthals. New York: The Experiment. ISBN 979-8-89303-070-9.
External links
- The Denisova Consortium's raw sequence data and alignments
- Human Timeline (Interactive) – Smithsonian, National Museum of Natural History (August 2016).
- Picture of Denisovan molar.
- Chris Stringer discusses the Harbin cranium
- Chris Stringer discusses the 1 million year old Yunxian cranium
Wikidata ☰ {{{from}}} entry
 |