Medicine:Medical open network for AI
 | |
| Developer(s) | NVIDIA Corporation, National Institutes of Health, King's College London |
|---|---|
| Initial release | Version 0.2.0 (November 23, 2021) |
| Stable release | Version 1.2.0 (April 30, 2023)
|
| Written in | Python |
| Platform | Cross-platform |
| Available in | English |
| Type | Health software |
| License | Apache Licence |
| Website | https://monai.io/ |
Medical open network for AI (MONAI) is an open-source, community-supported framework for Deep learning (DL) in healthcare imaging. MONAI provides a collection of domain-optimized implementations of various DL algorithms and utilities specifically designed for medical imaging tasks. MONAI is used in research and industry, aiding the development of various medical imaging applications, including image segmentation, image classification, image registration, and image generation.[1]
MONAI was first introduced in 2019 by a collaborative effort of engineers from NVIDIA, the National Institutes of Health, and the King's College London academic community. The framework was developed to address the specific challenges and requirements of DL applied to medical imaging.[1]
Built on top of PyTorch, a popular DL library, MONAI offers a high-level interface for performing everyday medical imaging tasks, including image preprocessing, augmentation, DL model training, evaluation, and inference for diverse medical imaging applications. MONAI simplifies the development of DL models for medical image analysis by providing a range of pre-built components and modules.[1][2][3]
Medical image analysis foundations

Medical imaging is a range of imaging techniques and technologies that enables clinicians to visualize the internal structures of the human body. It aids in diagnosing, treating, and monitoring various medical conditions, thus allowing healthcare professionals to obtain detailed and non-invasive images of organs, tissues, and physiological processes.[4]
Medical imaging has evolved, driven by technological advancements and scientific understanding. Today, it encompasses modalities such as X-ray, Computed Tomography (CT), Magnetic Resonance Imaging (MRI), ultrasound, nuclear medicine, and digital pathology, each offering capabilities and insights into human anatomy and pathology.[4]
The images produced by these medical imaging modalities are interpreted by radiologists, trained specialists in analyzing and diagnosing medical conditions based on the visual information captured in the images. In recent years, the field has witnessed advancements in computer-aided diagnosis, integrating Artificial intelligence and Deep learning techniques to automatize medical image analysis and assist radiologists in detecting abnormalities and improving diagnostic accuracy.[5]
Features
MONAI provides a robust suite of libraries, tools, and Software Development Kits (SDKs) that encompass the entire process of building medical imaging applications. It offers a comprehensive range of resources to support every stage of developing Artificial intelligence (AI) solutions in the field of medical imaging, from initial annotation (MONAI Label),[2] through models development and evaluation (MONAI Core),[1] and final application deployment (MONAI deploy application SDK).[3]
Medical data labeling

MONAI Label is a versatile tool that enhances the image labeling and learning process by incorporating AI assistance. It simplifies the task of annotating new datasets by leveraging AI algorithms and user interactions. Through this collaboration, MONAI Label trains an AI model for a specific task and continually improves its performance as it receives additional annotated images. The tool offers a range of features and integrations that streamline the annotation workflow and ensure seamless integration with existing medical imaging platforms.[6]
- AI-assisted annotation: MONAI Label assists researchers and practitioners in medical imaging by suggesting annotations based on user interactions by utilizing AI algorithms. This AI assistance significantly reduces the time and effort required for labeling new datasets, allowing users to focus on more complex tasks. The suggestions provided by MONAI Label enhance efficiency and accuracy in the annotation process.[6]
- Continuous learning: as users provide additional annotated images, MONAI Label utilizes this data to improve its performance over time. The tool updates its AI model with the newly acquired annotations, enhancing its ability to label images and adapt to specific tasks.[6]
- Integration with medical imaging platforms: MONAI Label integrates with medical imaging platforms such as 3D Slicer, Open Health Imaging Foundation viewer for radiology,[7] QuPath,[8] and digital slide archive for pathology.[9] These integrations enable communication between MONAI Label and existing medical imaging tools, facilitating collaborative workflows and ensuring compatibility with established platforms.[6]
- Custom viewer integration: developers have the flexibility to integrate MONAI Label into their custom image viewers using the provided server and client APIs. These APIs are abstracted and thoroughly documented, facilitating smooth integration with bespoke applications.[6]
Deep learning model development and evaluation
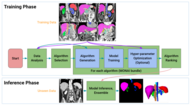
Within MONAI Core, researchers can find a collection of tools and functionalities for dataset processing, loading, Deep learning (DL) model implementation, and evaluation. These utilities allow researchers to evaluate the performance of their models. MONAI Core offers customizable training pipelines, enabling users to construct and train models that support various learning approaches such as supervised, semi-supervised, and self-supervised learning. Additionally, users have the flexibility to implement different computing strategies to optimize the training process.[1]
- Image I/O, processing, and augmentation: domain-specific APIs are available to transform data into arrays and different dictionary formats. Additionally, patch sampling strategies enable the generation of class-balanced samples from high-dimensional images. This ensures that the sampling process maintains balance and fairness across different classes present in the data. Furthermore, invertible transforms provided by MONAI Core allow for the reversal of model outputs to a previous preprocessing step. This is achieved by leveraging tracked metadata and applied operations, enabling researchers to interpret and analyze model results in the context of the original data.[1]
- Datasets and data loading: multi-threaded cache-based datasets support high-frequency data loading, public dataset availability accelerates model deployment and performance reproducibility, and custom APIs support compressed, image- and patched, and multimodal data sources.[1]
- Differentiable components, networks, losses,[10] and optimizers:[11] MONAI Core provides network layers and blocks that can seamlessly handle spatial 1D, 2D, and 3D inputs. Users have the flexibility to effortlessly integrate these layers, blocks, and networks into their personalized pipelines. The library also includes commonly used loss functions, such as Dice loss, Tversky loss, and Dice focal loss,[12] which have been (re-)implemented from literature. In addition, MONAI Core offers numerical optimization techniques like Novograd and utilities like learning rate finder to facilitate the optimization process.[13][1]
- Evaluation: MONAI Core provides a comprehensive set of evaluation metrics for assessing the performance of medical image models. These metrics include mean Dice, Receiving operating characteristic curves, Confusion matrices, Hausdorff distance, surface distance, and occlusion sensitivity.[14] The metric summary report generates statistical information such as mean, median, maximum, minimum, percentile, and standard deviation for the computed evaluation metrics.[14][1]
- GPU acceleration, performance profiling, and optimization: MONAI leverages a range of tools including DLProf,[15] Nsight,[16] NVTX,[17] and NVML[18] to detect performance bottlenecks. The distributed data-parallel APIs seamlessly integrate with the native PyTorch distributed module, PyTorch-ignite[19] distributed module, Horovod, XLA,[20] and the SLURM platform.[21]
- DL model collection: by offering the MONAI Model Zoo,[22] MONAI establishes itself as a platform that enables researchers and data scientists to access and share cutting-edge models developed by the community. Leveraging the MONAI Bundle format, users can seamlessly and efficiently utilize any model within the MONAI frameworks (Core, Label, or Deploy).[23]
AI-inference application development kit

The MONAI deploy application SDK offers a systematic series of steps empowering users to develop and fine-tune their AI models and workflows for deployment in clinical settings. These steps act as checkpoints, guaranteeing that the AI inference infrastructure adheres to the essential standards and requirements for seamless clinical integration.[3]
Key components of the MONAI Deploy Application SDK include:
- Pythonic framework for app development: the SDK presents a Python-based framework designed specifically for creating healthcare-focused applications. With its adaptable foundation, this framework enables the streamlined development of AI-driven applications tailored to the healthcare domain.[3]
- MONAI application package packaging mechanism: the SDK incorporates a tool for packaging applications into MONAI Application Packages[24] (MAP). These MAP instances establish a standardized format for bundling and deploying applications, ensuring portability and facilitating seamless distribution.[25]
- Local MAP execution via app runner: the SDK provides an app runner feature that enables the local execution of MAP instances. This functionality empowers developers to run and test their applications within a controlled environment, allowing prototyping and debugging.[26]
- Sample applications: the SDK includes a selection of sample applications that serve as both practical examples and starting points for developers. These sample applications showcase different use cases and exemplify best practices for effectively utilizing the MONAI Deploy framework.[3]
- API documentation: the SDK is complemented by comprehensive documentation that outlines the available APIs and provides guidance to developers on effectively leveraging the provided tools and functionalities.[3]
Applications
MONAI has found applications in various research studies and industry implementations across different anatomical regions. For instance, it has been utilized in academic research involving automatic cranio-facial implant design,[27] brain tumor analysis from Magnetic Resonance images,[28] identification of features in focal liver lesions from MRI scans,[29] radiotherapy planning for prostate cancer,[30] preparation of datasets for fluorescence microscopy imaging,[31] and classification of pulmonary nodules in lung cancer.[32]
In healthcare settings, hospitals have leveraged MONAI to enhance mammography reading by employing Deep learning models for breast density analysis. This approach reduce the waiting time for patients, allowing them to receive mammography results within 15 minutes. Consequently, clinicians save time, and patients experience shorter wait times. This advancement enables patients to engage in immediate discussions with their clinicians during the same appointment, facilitating prompt decision-making and discussion of next steps before leaving the facility. Moreover, hospitals can employ MONAI to identify indications of a COVID-19 patient's deteriorating condition or determine if they can be safely discharged, optimizing patient care and post-COVID-19 decision-making.[33]
In the corporate realm, companies choose MONAI to develop product applications addressing various clinical challenges. These include ultrasound-based scoliosis assessment, Artificial intelligence-based pathology image labeling, in-field pneumothorax detection using ultrasound, characterization of brain morphology, detection of micro-fractures in teeth, and non-invasive estimation of intracranial pressure.[34]
See also
- Artificial intelligence in healthcare
- Medical imaging
- Deep learning
- Image segmentation
- Image registration
- Image generation
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Cardoso, M. Jorge; Li, Wenqi; Brown, Richard (2022-10-04). "MONAI: An open-source framework for deep learning in healthcare". pp. 1–25. arXiv:2211.02701 [cs.LG].
- ↑ 2.0 2.1 Diaz-Pinto, Andres; Alle, Sachidanand; Nath, Vishwesh (2023-04-23). "MONAI Label: A framework for AI-assisted Interactive Labeling of 3D Medical Images". pp. 1–20. arXiv:2203.12362 [cs.HC].
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 MONAI Deploy App SDK, Project MONAI, 2023-06-29, https://github.com/Project-MONAI/monai-deploy-app-sdk, retrieved 2023-07-06
- ↑ 4.0 4.1 Dhawan, Atam P. (2011-01-24) (in en). Medical Image Analysis (1 ed.). Wiley. pp. 23–368. doi:10.1002/9780470918548. ISBN 978-0-470-62205-6. https://onlinelibrary.wiley.com/doi/book/10.1002/9780470918548.
- ↑ Doi, Kunio (2007). "Computer-aided diagnosis in medical imaging: Historical review, current status and future potential" (in en). Computerized Medical Imaging and Graphics 31 (4–5): 198–211. doi:10.1016/j.compmedimag.2007.02.002. PMID 17349778.
- ↑ 6.0 6.1 6.2 6.3 6.4 MONAI Label, Project MONAI, 2023-07-06, https://github.com/Project-MONAI/MONAILabel, retrieved 2023-07-06
- ↑ "OHIF Viewer". https://viewer.ohif.org/.
- ↑ "QuPath" (in en). https://qupath.github.io/.
- ↑ "Harness the full potential of your digital pathology data". https://myawesomeurl.com//digital_slide_archive/.
- ↑ "Descending into ML: Training and Loss | Machine Learning" (in en). https://developers.google.com/machine-learning/crash-course/descending-into-ml/training-and-loss.
- ↑ An, Li-Bao (2011). "Cutting parameter optimization in milling operations by various solution methods". 2011 International Conference on Machine Learning and Cybernetics. IEEE. pp. 422–427. doi:10.1109/icmlc.2011.6016679. ISBN 978-1-4577-0305-8.
- ↑ "Loss functions — MONAI 1.2.0 Documentation". https://docs.monai.io/en/stable/losses.html.
- ↑ "Optimizers — MONAI 1.2.0 Documentation". https://docs.monai.io/en/stable/optimizers.html.
- ↑ 14.0 14.1 "Metrics — MONAI 1.2.0 Documentation". https://docs.monai.io/en/stable/metrics.html.
- ↑ "DLProf User Guide". https://docs.nvidia.com/deeplearning/frameworks/pdf/DLProf-User-Guide.pdf.
- ↑ "Nsight Systems" (in en). 2018-03-12. https://developer.nvidia.com/nsight-systems.
- ↑ "NVIDIA Tools Extension (NVTX) Documentation". https://docs.nvidia.com/nvtx/.
- ↑ "NVML API Reference Guide :: GPU Deployment and Management Documentation" (in en-us). http://docs.nvidia.com/deploy/nvml-api/index.html.
- ↑ "PyTorch-Ignite" (in en-us). https://pytorch-ignite.ai/.
- ↑ "XLA: Optimizing Compiler for Machine Learning" (in en). https://www.tensorflow.org/xla.
- ↑ "Slurm Workload Manager - Documentation". https://slurm.schedmd.com/documentation.html.
- ↑ "MONAI Model Zoo" (in en). https://monai.io/model-zoo.html.
- ↑ "Model Bundle — MONAI 1.2.0 Documentation". https://docs.monai.io/en/stable/bundle.html.
- ↑ "Deploying and Hosting MONAI App Package — MONAI Deploy App SDK 0.4.0 Documentation". https://docs.monai.io/projects/monai-deploy-app-sdk/en/latest/developing_with_sdk/deploying_and_hosting_map.html.
- ↑ "Packaging app — MONAI Deploy App SDK 0.4.0 Documentation". https://docs.monai.io/projects/monai-deploy-app-sdk/en/latest/developing_with_sdk/packaging_app.html.
- ↑ "Executing packaged app locally — MONAI Deploy App SDK 0.4.0 Documentation". https://docs.monai.io/projects/monai-deploy-app-sdk/en/latest/developing_with_sdk/executing_packaged_app_locally.html.
- ↑ Li, Jianning; Ferreira, André; Puladi, Behrus; Alves, Victor; Kamp, Michael; Kim, Moon; Nensa, Felix; Kleesiek, Jens et al. (2023). "Open-source skull reconstruction with MONAI". SoftwareX 23: 101432. doi:10.1016/j.softx.2023.101432. ISSN 2352-7110. Bibcode: 2023SoftX..2301432L.
- ↑ Sharma, Suraj Prakash; Sampath, Nalini (2022-06-24). "Data Augmentation for Brain Tumor Segmentation using MONAI Framework". 2022 2nd International Conference on Intelligent Technologies (CONIT). IEEE. pp. 1–8. doi:10.1109/conit55038.2022.9847822. ISBN 978-1-6654-8407-7. http://dx.doi.org/10.1109/conit55038.2022.9847822.
- ↑ Stollmayer, Róbert; Budai, Bettina Katalin; Rónaszéki, Aladár; Zsombor, Zita; Kalina, Ildikó; Hartmann, Erika; Tóth, Gábor; Szoldán, Péter et al. (2022-05-05). "Focal Liver Lesion MRI Feature Identification Using Efficientnet and MONAI: A Feasibility Study". Cells 11 (9): 1558. doi:10.3390/cells11091558. ISSN 2073-4409. PMID 35563862.
- ↑ Belue, Mason J.; Harmon, Stephanie A.; Patel, Krishnan; Daryanani, Asha; Yilmaz, Enis Cagatay; Pinto, Peter A.; Wood, Bradford J.; Citrin, Deborah E. et al. (2022). "Development of a 3D CNN-based AI Model for Automated Segmentation of the Prostatic Urethra". Academic Radiology 29 (9): 1404–1412. doi:10.1016/j.acra.2022.01.009. ISSN 1076-6332. PMID 35183438. PMC 9339453. http://dx.doi.org/10.1016/j.acra.2022.01.009.
- ↑ Poon, Charissa; Teikari, Petteri; Rachmadi, Muhammad Febrian; Skibbe, Henrik; Hynynen, Kullervo (2022-07-20). MiniVess: A dataset of rodent cerebrovasculature from in vivo multiphoton fluorescence microscopy imaging. pp. 1–11. doi:10.1101/2022.07.19.500542. http://dx.doi.org/10.1101/2022.07.19.500542. Retrieved 2023-07-06.
- ↑ Kaliyugarasan, Satheshkumar; Lundervold, Arvid; Lundervold, Alexander Selvikvåg (2021). "Pulmonary Nodule Classification in Lung Cancer from 3D Thoracic CT Scans Using fastai and MONAI". International Journal of Interactive Multimedia and Artificial Intelligence 6 (7): 83. doi:10.9781/ijimai.2021.05.002. ISSN 1989-1660.
- ↑ Durlach, Peter (2023-05-11). "Microsoft and our partners: Bringing AI models to clinical settings" (in en-US). https://www.microsoft.com/en-us/industry/blog/healthcare/2023/05/11/bringing-medical-imaging-ai-models-directly-into-clinical-settings-at-scale/.
- ↑ Dennison, Jaisil Rose (2022-08-11). "Leveraging MONAI for Medical AI" (in en-US). https://www.kitware.com/leveraging-monai-for-medical-ai/.
Further reading
- Diaz-Pinto, Andres; Mehta, Pritesh; Alle, Sachidanand; Asad, Muhammad; Brown, Richard; Nath, Vishwesh; Ihsani, Alvin; Antonelli, Michela et al. (2022), "DeepEdit: Deep Editable Learning for Interactive Segmentation of 3D Medical Images", Data Augmentation, Labelling, and Imperfections, Lecture Notes in Computer Science, 13567, Cham: Springer Nature Switzerland, pp. 11–21, doi:10.1007/978-3-031-17027-0_2, ISBN 978-3-031-17026-3, http://dx.doi.org/10.1007/978-3-031-17027-0_2, retrieved 2023-07-06
- Hatamizadeh, Ali; Tang, Yucheng; Nath, Vishwesh; Yang, Dong; Myronenko, Andriy; Landman, Bennett; Roth, Holger R.; Xu, Daguang (2022). "UNETR: Transformers for 3D Medical Image Segmentation". 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV). IEEE. pp. 574–584. doi:10.1109/wacv51458.2022.00181. ISBN 978-1-6654-0915-5. http://dx.doi.org/10.1109/wacv51458.2022.00181.
- Hatamizadeh, Ali; Nath, Vishwesh; Tang, Yucheng; Yang, Dong; Roth, Holger R.; Xu, Daguang (2022). "Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images". in Crimi, Alessandro; Bakas, Spyridon (in en). Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Lecture Notes in Computer Science. 12962. Cham: Springer International Publishing. pp. 272–284. doi:10.1007/978-3-031-08999-2_22. ISBN 978-3-031-08999-2. https://link.springer.com/chapter/10.1007/978-3-031-08999-2_22.
External links
