Chemistry:Biodesulfurization
Biodesulfurization is the process of removing sulfur from crude oil through the use of microorganisms or their enzymes.[1]
Background
Crude oil contains sulfur in its composition, with the latter being the most abundant element after carbon and hydrogen.[2] Depending on its source, the amount of sulfur present in crude oil can range from 0.05 to 10%.[3] Accordingly, the oil can be classified as sweet or sour if the sulfur concentration is below or above 0.5%, respectively.[4]
The combustion of crude oil releases sulfur oxides (SOx) to the atmosphere, which are harmful to public health and contribute to serious environmental effects such as air pollution and acid rains.[2][3] In addition, the sulfur content in crude oil is a major problem for refineries, as it promotes the corrosion of the equipment and the poisoning of the noble metal catalysts.[5] The levels of sulfur in any oil field are too high for the fossil fuels derived from it (such as gasoline, diesel, or jet fuel ) to be used in combustion engines without pre-treatment to remove organosulfur compounds.
The reduction of the concentration of sulfur in crude oil becomes necessary to mitigate one of the leading sources of the harmful health and environmental effects caused by its combustion. In this sense, the European union has taken steps to decrease the sulfur content in diesel below 10 ppm,[6] while the US has made efforts to restrict the sulfur content in diesel and gasoline to a maximum of 15 ppm.[7] The reduction of sulfur compounds in oil fuels can be achieved by a process named desulfurization.[8] Methods used for desulfurization include, among others, hydrodesulfurization,[9] oxidative desulfurization,[10] extractive desulfurization,[11] and extraction by ionic liquids.[12]
Despite their efficiency at reducing sulfur content, the conventional desulfurization methods are still accountable for a significant amount of the CO2 emissions associated with the crude oil refining process, releasing up to 9000 metric tons per year.[13] Furthermore, these processes usually require large amounts of energy, and are accompanied by massive costs for the industries that employ them. A greener and also complementary alternative process to the conventional desulfurization methods is biodesulfurization.[3]
Biodesulfurization implementation and pathways
It has been observed that there are sulfur-dependent bacteria that make use of the sulfur in sulfur-containing compounds in their life cycles (either in their growth or metabolic processes), producing molecules with lower/no content in sulfur.[14] In particular, heteroaromatic compounds, namely thiophenes and their derivatives, were observed to constitute important substrates for bacteria.[15][16]
Biodesulfurization is an attractive alternative to sulfur removal, particularly in the crude oil fractions where there is an abundance of sulfur heterocycles.[17][3] To date, pilot attempts for industrial applications have resorted to the use of whole bacterial systems, because biodesulfurization involves a sequential cascade of reactions by different enzymes and a large amount of cofactors participating in redox reactions either with the sulfur atom or molecular oxygen.[18] However, they lacked the scalability desired for an industrial setup due to overall low enzyme efficiency, product feedback inhibition mechanisms and toxicity, or inadequate conditions for long-term bacterial growth.[14] While cell-free recombinant enzymes would be desirable, known implementations are still well below the efficiency met for whole-cell ones.[19]
There are two main pathways through which bacteria remove sulfur from sulfur-containing compounds: ring destructive pathways and sulfur-specific pathways. The ring destructive pathway consists of the selective cleavage of carbon-carbon bonds with release of small organic sulfides soluble in the surrounding aqueous environment, whereas the sulfur-specific pathways rely on successive sulfur redox reactions to release sulfur either as sulfide or sulfite anions as byproducts.[3] The latter have thus been considered as a very promising pathway to produce sulfur-free compounds with a high calorific content, in particular in the desulfurization of sulfur heterocycles abundant in sour crude oil fractions.[1]
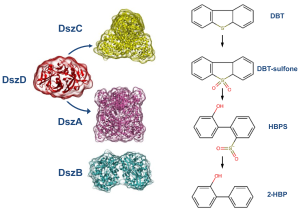
The most studied ring destructive pathway is the Kodama pathway and it was initially identified in Pseudomonas abikonensis and Pseudomonas jijani.[21] The pathway comprises four main steps: i) the successive hydroxylation by NADH-dependent dioxygenases of the carbons in one of the aromatic rings, followed by ii) the dehydrogenation of the ring by a NAD+ cofactor and further iii) oxygenation promoting ring cleavage and formation of a pyruvyl branch; concluding with iv) the hydrolysis of the pyruvyl substituent to release pyruvate and the remaining of the substrate.[20][22] Since the end products of the pathway are still water soluble sulfur compounds, the pathway has often been disregarded as an appealing pathway for industrial applications, in particular by the oil industry.[23] The most well-studied sulfur specific pathway is the 4S pathway, first discovered in the bacterium Rhodococcus erythropolis (strain IGTS8),[1] which was observed to remove sulfur from dibenzothiophenes and derivatives in three steps: i) a double oxidation of the sulfur (to sulfoxide and sulfone) performed by a flavin-dependent monoxygenase, followed by ii) a carbon-sulfur bond cleavage by a second flavin-dependent monoxygenase and a iii) desulfination reaction through which 2-hydroxybiphenyl and sulfite are produced.[24] In total, four enzymes are required for the process: three of which are encoded in the dszABC genes (the flavin-dependent monoxygenases DszA and DszC, and the desulfinase DszB) and fourth chromosome encoded enzyme, DszD, which is responsible for the regeneration and supply of the flavin mononucleotide cofactor required for DszA and DszC.[25][26]
It has also been observed that some anaerobic bacteria can use an alternative sulfur-specific pathway to produce hydrogen sulfide instead.[27] However, to date, the desulfurization of fractions such as bitumen, vacuum gas oil, or deasphalted oil has not been observed[28]
The aerobic 4S pathway
The 4S pathway is a sulfur-specific metabolic pathway of oxidative desulfurization that converts dibenzothiophene (DBT) into 2-hydroxybiphenyl and sulfite. It uses a total of four NADH molecules (three required by DszD to generate FMNH2 and a fourth to regenerate the FMN-oxide byproduct of DszA) and three molecules of oxygen, thus producing NAD+ and water as byproducts.[26]
DszC is the first enzyme to intervene in the pathway in two sequential steps, catalyzing the double oxidation of DBT first into DBT-sulfoxide and then into DBT-sulfone. It requires FMNH2 as cofactor, which is supplied by DszD, and molecular oxygen. For that reason, the efficiency of this enzyme is dependent on the activity of DszD and on environmental oxygenation. The reaction catalyzed by DszC involves three phases: 1) molecular oxygen activation leading to the formation of a hydroperoxyflavin-intermediate (C4aOOH); 2) oxidation of DBT to DBTO; and 3) dehydration of FMN.[29] DszC is the second least efficient enzyme in the pathway with a particularly low kcat of 1.6 ± 0.3 min−1.[24] It is also severely affected from feedback inhibition caused mostly by HPBS and 2-HBP, the products of DszA and DszB respectively,[24] For that reason, it has been targeted for optimization through enzyme engineering.
DszA is responsible for the third step of the pathway. It catalyzes the first carbon-sulfur bond cleavage, converting DBT-sulfone into 2-hydroxybiphenyl-2-sulfinate. Like DszC, DszA also requires FMNH2 provided by DszD and molecular oxygen for its catalytic cycle. Nonetheless, the reaction rate of DszA is about seven times faster than DszC. However, like DszC, it suffers feedback inhibition by the final product of the pathway, 2-HBP.
At last, the desulfinase (DszB) cleaves the remaining carbon-sulfur bond in 2-hydroxybiphenyl-2-sulfinate converting it into the sulfur-free 2-hydroxybiphenyl in a two step mechanism. In the first, and rate-limiting, step, 2-hydroxybiphenyl-2-sulfinate is protonated by Cys27 in its electrophilic carbon leading to the cleavage of the carbon-sulfur bond and displacement of SO2. In the second step, a water molecule is deprotonated by Cys27 followed by the hydroxide attack to SO2 forming HSO3-.[30] DszB is the least efficient enzyme on the pathway making it an appealing target for enhancement through protein engineering.[24]
The NADH-FMN oxidoreductase (DszD) regenerates the FMNH2 cofactor needed for the reactions catalyzed by DszC and DszA, through the oxidation of NADH to NAD+ in a two step mechanism. The first step corresponds to a hydride transfer from the nicotinamide moiety of NADH to the central nitrogen in the isoalloxazine moiety of the oxidized FMN forming FMNH. In the second step, a water molecule protonates the N1 atom of FMNH giving FMNH2.[31]
Engineering of 4S pathway enzymes
The desulfurization rate for the wild-type 4S pathway enzymes is low when compared to the rate that needs to be achieved for a viable application in the industrial sector. An increase of 500-fold on the overall rate of the pathway is the required improvement for an efficient application of this biodesulfurization method.[32]
Directed evolution, rational design or a combination of both strategies are some of the approaches that have been applied to tackle the lack of catalytic efficiency and stability of the 4S enzymes. The 4S pathway best improvement to date was obtained by a directed evolution approach in which Rhodococcus strains were transformed with a plasmid encoding a modified dsz operon (which encodes for DszA, DszB and DszC). After 40 subculturing events in a medium in which DBT was the sole sulfur source, the modified Rhodococcus strains presented a 35-fold improvement.[33]
The strong feedback inhibition of DszC was also tackled by a combination of directed evolution and rational design approach to desensitize DszC to the 4S pathway product, HBP. The bacterial strain expressing the DszC A101K mutant showed higher activity relative to the wild-type strain. Additionally docking of HBP to the protein revealed that HBP forms a π-interaction with Trp327, thus inhibiting DszC. The A101K/W327C (AKWC) double mutant revealed to be desensitized to low HBP concentrations and the bacterial strain expressing the AKWC DszC was 14-fold more efficient than the wild-type strain.[34]
DszB, the final enzyme in the pathway, is also one of the slowest with a turnover rate of 1.7 ± 0.2 min−1, becoming a major bottleneck of the 4S pathway. A computational rational design approach determined a set of mutations that could accelerate the charge transfer occurring in the active site during DszB reaction mechanism, reducing the activation energy for the reaction and potentially increasing its turnover rate.[30]
DszB's catalytic efficiency and thermostability was also addressed in an experimental mutagenesis approach, the Y63F/Q65H double mutant revealed an increase in the enzyme's thermostability without loss of catalytic efficiency.[35]
DszD has also been targeted for rate enhancing mutation on the Thr62 residue. Mutation of Thr62 by Asn and Ala residues managed to increase its activity 5- and 7-fold, respectively.[36]
A computational study demonstrated that substitutions in position 62 of DszD sequence have a major impact in the activation energy for the hydride transfer reaction from NADH to FAD. The Thr62 mutation by an Asp residue returns the lowest activation energy from all possible mutants at this position due to the stabilization effect induced by Asp negative charge.[37]
See also
References
- ↑ Jump up to: 1.0 1.1 1.2 Borgne, Sylvie Le; Quintero, Rodolfo (May 2003). "Biotechnological processes for the refining of petroleum" (in en). Fuel Processing Technology 81 (2): 155–169. doi:10.1016/S0378-3820(03)00007-9. https://linkinghub.elsevier.com/retrieve/pii/S0378382003000079.
- ↑ Jump up to: 2.0 2.1 Demirbas, A.; Alidrisi, H.; Balubaid, M. A. (January 2, 2015). "API Gravity, Sulfur Content, and Desulfurization of Crude Oil" (in en). Petroleum Science and Technology 33 (1): 93–101. doi:10.1080/10916466.2014.950383. ISSN 1091-6466. http://www.tandfonline.com/doi/abs/10.1080/10916466.2014.950383.
- ↑ Jump up to: 3.0 3.1 3.2 3.3 3.4 Mohebali, Ghasemali; Ball, Andrew S. (May 2016). "Biodesulfurization of diesel fuels – Past, present and future perspectives" (in en). International Biodeterioration & Biodegradation 110: 163–180. doi:10.1016/j.ibiod.2016.03.011. https://linkinghub.elsevier.com/retrieve/pii/S0964830516300804.
- ↑ "An Introduction to Petroleum Refining and the Production of Ultra Low Sulfur Gasoline and Diesel Fuel" (in en-US). https://theicct.org/publication/an-introduction-to-petroleum-refining-and-the-production-of-ultra-low-sulfur-gasoline-and-diesel-fuel/.
- ↑ Kilbane, J.J.; Le Borgne, S. (2004), "Chapter 2 Petroleum biorefining: the selective removal of sulfur, nitrogen, and metals" (in en), Studies in Surface Science and Catalysis (Elsevier) 151: pp. 29–65, doi:10.1016/s0167-2991(04)80143-5, ISBN 978-0-444-51699-2, https://linkinghub.elsevier.com/retrieve/pii/S0167299104801435, retrieved 2022-12-07
- ↑ (in en) Directive 2009/30/EC of the European Parliament and of the Council of 23 April 2009 amending Directive 98/70/EC as regards the specification of petrol, diesel and gas-oil and introducing a mechanism to monitor and reduce greenhouse gas emissions and amending Council Directive 1999/32/EC as regards the specification of fuel used by inland waterway vessels and repealing Directive 93/12/EEC (Text with EEA relevance), OJ L, June 5, 2009, http://data.europa.eu/eli/dir/2009/30/oj/eng, retrieved 2022-12-07
- ↑ US EPA, OAR (April 10, 2015). "Diesel Fuel Standards and Rulemakings" (in en). https://www.epa.gov/diesel-fuel-standards/diesel-fuel-standards-and-rulemakings.
- ↑ Babich, I (April 2003). "Science and technology of novel processes for deep desulfurization of oil refinery streams: a review⋆" (in en). Fuel 82 (6): 607–631. doi:10.1016/S0016-2361(02)00324-1. https://linkinghub.elsevier.com/retrieve/pii/S0016236102003241.
- ↑ Babich, I. V; Moulijn, J. A (April 1, 2003). "Science and technology of novel processes for deep desulfurization of oil refinery streams: a review☆" (in en). Fuel 82 (6): 607–631. doi:10.1016/S0016-2361(02)00324-1. ISSN 0016-2361. https://www.sciencedirect.com/science/article/pii/S0016236102003241.
- ↑ Campos-Martin, J.M.; Capel-Sanchez, M.C.; Perez-Presas, P.; Fierro, J.L.G. (March 9, 2010). "Oxidative processes of desulfurization of liquid fuels" (in en). Journal of Chemical Technology & Biotechnology 85 (7): 879–890. doi:10.1002/jctb.2371. https://onlinelibrary.wiley.com/doi/10.1002/jctb.2371.
- ↑ Javadli, Rashad; de Klerk, Arno (March 2012). "Desulfurization of heavy oil" (in en). Applied Petrochemical Research 1 (1–4): 3–19. doi:10.1007/s13203-012-0006-6. ISSN 2190-5525.
- ↑ Hosseini, Alireza; Khoshsima, Ali; Sabzi, Mazaher; Rostam, Ata (April 21, 2022). "Toward Application of Ionic Liquids to Desulfurization of Fuels: A Review" (in en). Energy & Fuels 36 (8): 4119–4152. doi:10.1021/acs.energyfuels.1c03974. ISSN 0887-0624. https://pubs.acs.org/doi/10.1021/acs.energyfuels.1c03974.
- ↑ Miranda-Galindo, Erick Yair; Segovia-Hernández, Juan Gabriel; Hernández, Salvador; Bonilla-Petriciolet, Adrián (October 22, 2014). "Multiobjective Optimization of a Hydrodesulfurization Process of Diesel Using Distillation with Side Reactor" (in en). Industrial & Engineering Chemistry Research 53 (42): 16425–16435. doi:10.1021/ie501940v. ISSN 0888-5885.
- ↑ Jump up to: 14.0 14.1 Boniek, Douglas; Figueiredo, Débora; dos Santos, Antônio Fernando Batista; de Resende Stoianoff, Maria Aparecida (January 2015). "Biodesulfurization: a mini review about the immediate search for the future technology" (in en). Clean Technologies and Environmental Policy 17 (1): 29–37. doi:10.1007/s10098-014-0812-x. ISSN 1618-954X. http://link.springer.com/10.1007/s10098-014-0812-x.
- ↑ Gunam, Ida Bagus Wayan; Yaku, Yosuke; Hirano, Makoto; Yamamura, Kenta; Tomita, Fusao; Sone, Teruo; Asano, Kozo (April 2006). "Biodesulfurization of alkylated forms of dibenzothiophene and benzothiophene by Sphingomonas subarctica T7b" (in en). Journal of Bioscience and Bioengineering 101 (4): 322–327. doi:10.1263/jbb.101.322. PMID 16716940. https://linkinghub.elsevier.com/retrieve/pii/S1389172306705886.
- ↑ Davoodi-Dehaghani, Fatemeh; Vosoughi, Manouchehr; Ziaee, Abed Ali (February 2010). "Biodesulfurization of dibenzothiophene by a newly isolated Rhodococcus erythropolis strain" (in en). Bioresource Technology 101 (3): 1102–1105. doi:10.1016/j.biortech.2009.08.058. PMID 19819129. https://linkinghub.elsevier.com/retrieve/pii/S0960852409010335.
- ↑ Jump up to: 17.0 17.1 17.2 Sousa, João P. M.; Ferreira, Pedro; Neves, Rui P. P.; Ramos, Maria J.; Fernandes, Pedro A. (2020). "The bacterial 4S pathway – an economical alternative for crude oil desulphurization that r educes CO2 emissions" (in en). Green Chemistry 22 (22): 7604–7621. doi:10.1039/D0GC02055A. ISSN 1463-9262. http://xlink.rsc.org/?DOI=D0GC02055A.
- ↑ Kilbane, J.J.; Le Borgne, S. (2004), "Chapter 2 Petroleum biorefining: the selective removal of sulfur, nitrogen, and metals" (in en), Studies in Surface Science and Catalysis (Elsevier) 151: pp. 29–65, doi:10.1016/s0167-2991(04)80143-5, ISBN 978-0-444-51699-2, https://linkinghub.elsevier.com/retrieve/pii/S0167299104801435, retrieved 2022-12-08
- ↑ Setti, L.; Lanzarini, G.; Pifferi, P.G. (November 1997). "Whole cell biocatalysis for an oil desulfurization process" (in en). Fuel Processing Technology 52 (1–3): 145–153. doi:10.1016/S0378-3820(97)00023-4. https://linkinghub.elsevier.com/retrieve/pii/S0378382097000234.
- ↑ Jump up to: 20.0 20.1 Borgne, S. Le; Ayala, M. (2010), Timmis, Kenneth N., ed., "Microorganisms Utilizing Sulfur-Containing Hydrocarbons" (in en), Handbook of Hydrocarbon and Lipid Microbiology (Berlin, Heidelberg: Springer Berlin Heidelberg): pp. 2129–2141, doi:10.1007/978-3-540-77587-4_154, ISBN 978-3-540-77584-3, http://link.springer.com/10.1007/978-3-540-77587-4_154, retrieved 2022-12-08
- ↑ Kodama, Koki; Umehara, Kazuyoshi; Shimizu, Katsumi; Nakatani, Shigeru; Minoda, Yasuji; Yamada, Koichi (January 1973). "Identification of Microbial Products from Dibenzothiophene and Its Proposed Oxidation Pathway" (in en). Agricultural and Biological Chemistry 37 (1): 45–50. doi:10.1080/00021369.1973.10860640. ISSN 0002-1369. https://academic.oup.com/bbb/article/37/1/45-50/5979328.
- ↑ Seo, Jong-Su; Keum, Young-Soo; Li, Qing (January 13, 2009). "Bacterial Degradation of Aromatic Compounds" (in en). International Journal of Environmental Research and Public Health 6 (1): 278–309. doi:10.3390/ijerph6010278. ISSN 1660-4601. PMID 19440284.
- ↑ Mohebali, G.; Ball, A.S.; Rasekh, B.; Kaytash, A. (March 2007). "Biodesulfurization potential of a newly isolated bacterium, Gordonia alkanivorans RIPI90A" (in en). Enzyme and Microbial Technology 40 (4): 578–584. doi:10.1016/j.enzmictec.2006.05.012. https://linkinghub.elsevier.com/retrieve/pii/S0141022906002833.
- ↑ Jump up to: 24.0 24.1 24.2 24.3 Abin-Fuentes, Andres; Mohamed, Magdy El-Said; Wang, Daniel I. C.; Prather, Kristala L. J. (December 15, 2013). "Exploring the Mechanism of Biocatalyst Inhibition in Microbial Desulfurization" (in en). Applied and Environmental Microbiology 79 (24): 7807–7817. doi:10.1128/AEM.02696-13. ISSN 0099-2240. PMID 24096431. Bibcode: 2013ApEnM..79.7807A.
- ↑ Oldfield, Christopher; Wood, Nicola T.; Gilbert, Steven C.; Murray, Frazer D.; Faure, Fabrice R. (1998). "Desulphurisation of benzothiophene and dibenzothiophene by actinomycete organisms belonging to the genus Rhodococcus, and related taxa". Antonie van Leeuwenhoek 74 (1/3): 119–132. doi:10.1023/A:1001724516342. PMID 10068795. http://link.springer.com/10.1023/A:1001724516342.
- ↑ Jump up to: 26.0 26.1 Oldfield, Christopher; Pogrebinsky, Olga; Simmonds, Julie; Olson, Edwin S.; Kulpa, Charles F. (September 1, 1997). "Elucidation of the metabolic pathway for dibenzothiophene desulphurization by Rhodococcus sp. strain IGTS8 (ATCC 53968)" (in en). Microbiology 143 (9): 2961–2973. doi:10.1099/00221287-143-9-2961. ISSN 1350-0872. PMID 9308179.
- ↑ Bahrami, A.; Shojaosadati, S.A.; Mohebali, G. (2001). "Biodegradation of dibenzothiophene by thermophilic bacteria". Biotechnology Letters 23 (11): 899–901. doi:10.1023/A:1010592615572. http://link.springer.com/10.1023/A:1010592615572.
- ↑ Armstrong, Stephen M.; Sankey, Bruce M.; Voordouw, Gerrit (February 1997). "Evaluation of sulfate-reducing bacteria for desulfurizing bitumen or its fractions" (in en). Fuel 76 (3): 223–227. doi:10.1016/S0016-2361(96)00226-8. https://linkinghub.elsevier.com/retrieve/pii/S0016236196002268.
- ↑ Barbosa, Ana C. C.; Neves, Rui P. P.; Sousa, Sérgio F.; Ramos, Maria J.; Fernandes, Pedro A. (October 5, 2018). "Mechanistic Studies of a Flavin Monooxygenase: Sulfur Oxidation of Dibenzothiophenes by DszC" (in en). ACS Catalysis 8 (10): 9298–9311. doi:10.1021/acscatal.8b01877. ISSN 2155-5435. https://pubs.acs.org/doi/10.1021/acscatal.8b01877.
- ↑ Jump up to: 30.0 30.1 Sousa, João P. M.; Neves, Rui P. P.; Sousa, Sérgio F.; Ramos, Maria J.; Fernandes, Pedro A. (August 21, 2020). "Reaction Mechanism and Determinants for Efficient Catalysis by DszB, a Key Enzyme for Crude Oil Bio-desulfurization" (in en). ACS Catalysis 10 (16): 9545–9554. doi:10.1021/acscatal.0c03122. ISSN 2155-5435. https://pubs.acs.org/doi/10.1021/acscatal.0c03122.
- ↑ Sousa, Sérgio F.; Sousa, Joana F. M.; Barbosa, Ana C. C.; Ferreira, Cleide E.; Neves, Rui P. P.; Ribeiro, António J. M.; Fernandes, Pedro A.; Ramos, Maria João (July 14, 2016). "Improving the Biodesulfurization of Crude Oil and Derivatives: A QM/MM Investigation of the Catalytic Mechanism of NADH-FMN Oxidoreductase (DszD)" (in en). The Journal of Physical Chemistry A 120 (27): 5300–5306. doi:10.1021/acs.jpca.6b01536. ISSN 1089-5639. PMID 27128525. Bibcode: 2016JPCA..120.5300S. https://pubs.acs.org/doi/10.1021/acs.jpca.6b01536.
- ↑ Kilbane, John J (June 1, 2006). "Microbial biocatalyst developments to upgrade fossil fuels" (in en). Current Opinion in Biotechnology. Environmental biotechnology/Energy biotechnology 17 (3): 305–314. doi:10.1016/j.copbio.2006.04.005. ISSN 0958-1669. PMID 16678400. https://www.sciencedirect.com/science/article/pii/S0958166906000577.
- ↑ Pan, Jie; Wu, Fan; Wang, Jia; Yu, Linqing; Khayyat, Naghmeh Hassanzadeh; Stark, Benjamin C.; Kilbane, John J. (October 1, 2013). "Enhancement of desulfurization activity by enzymes of the Rhodococcus dsz operon through coexpression of a high sulfur peptide and directed evolution" (in en). Fuel 112: 385–390. doi:10.1016/j.fuel.2013.04.065. ISSN 0016-2361. https://www.sciencedirect.com/science/article/pii/S0016236113003669.
- ↑ Li, Lu; Liao, Yibo; Luo, Yifan; Zhang, Guangming; Liao, Xihao; Zhang, Wei; Zheng, Suiping; Han, Shuangyan et al. (June 21, 2019). "Improved Efficiency of the Desulfurization of Oil Sulfur Compounds in Escherichia coli Using a Combination of Desensitization Engineering and DszC Overexpression" (in en). ACS Synthetic Biology 8 (6): 1441–1451. doi:10.1021/acssynbio.9b00126. ISSN 2161-5063. PMID 31132321. https://pubs.acs.org/doi/10.1021/acssynbio.9b00126.
- ↑ OHSHIRO, Takashi; OHKITA, Ryo; TAKIKAWA, Takeshi; MANABE, Masanori; LEE, Woo Cheol; TANOKURA, Masaru; IZUMI, Yoshikazu (November 23, 2007). "Improvement of 2′-Hydroxybiphenyl-2-sulfinate Desulfinase, an Enzyme Involved in the Dibenzothiophene Desulfurization Pathway, fromRhodococcus erythropolisKA2-5-1 by Site-Directed Mutagenesis". Bioscience, Biotechnology, and Biochemistry 71 (11): 2815–2821. doi:10.1271/bbb.70436. ISSN 0916-8451. PMID 17986771.
- ↑ Kamali, Nasrin; Tavallaie, Mahmood; Bambai, Bijan; Karkhane, Ali Asghar; Miri, Mandana (July 1, 2010). "Site-directed mutagenesis enhances the activity of NADH-FMN oxidoreductase (DszD) activity of Rhodococcus erythropolis" (in en). Biotechnology Letters 32 (7): 921–927. doi:10.1007/s10529-010-0254-4. ISSN 1573-6776. PMID 20349330. https://doi.org/10.1007/s10529-010-0254-4.
- ↑ Ferreira, Pedro; Sousa, Sérgio F.; Fernandes, Pedro A.; Ramos, Maria João (December 6, 2017). "Improving the Catalytic Power of the DszD Enzyme for the Biodesulfurization of Crude Oil and Derivatives" (in en). Chemistry – A European Journal 23 (68): 17231–17241. doi:10.1002/chem.201704057. PMID 28976031. https://onlinelibrary.wiley.com/doi/10.1002/chem.201704057.
 |