Biology:15-Cis-phytoene desaturase
| 15-cis-phytoene desaturase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 1.3.5.5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
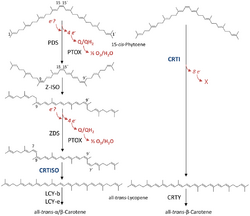
15-cis-phytoene desaturases (PDS, plant-type phytoene desaturases) (EC 1.3.5.5, 15-cis-phytoene:plastoquinone oxidoreductase), are enzymes involved in the carotenoid biosynthesis in plants and cyanobacteria.[2] Phytoene desaturases are membrane-bound enzymes localized in plastids and introduce two double bonds into their colorless substrate phytoene by dehydrogenation and isomerize two additional double bonds.[3][4] This reaction starts a biochemical pathway involving three further enzymes (zeta-carotene isomerase, zeta-carotene desaturase and carotene cis-trans isomerase) called the poly-cis pathway and leads to the red colored lycopene. The homologous phytoene desaturase found in bacteria and fungi (CrtI) converts phytoene directly to lycopene by an all-trans pathway.[5]
Biochemistry
PDS converts 15-cis-phytoene into 9,15,9'-tri-cis-ζ-carotene through reduction of the enzymes non-covalently bound FAD cofactor.[6] This conversion introduces two additional double bonds at positions 11 and 11' of the carbon chain and isomerizes two adjacent already existing double bonds at positions 9 and 9' from trans to cis. The electrons involved in the reaction are subsequently transferred onto plastoquinone[7] and to plastid terminal oxidase PTOX ultimately coupling the desaturation to oxygen reduction. Disruption of this biosynthesis step results in albinism and stunted plant growth.[8]
Applications
Disruption of PDS function can be achieved by bleaching herbicides such as norflurazon[9] and fluridone.[10] These inhibitors occupy the binding pocket of plastoquinone within the enzyme thus blocking it from its function.[1] Due to the clear effect of PDS disruption in plants, the corresponding gene was targeted to showcase successful genome editing in fruit such as apples,[11] grapes[12] or bananas[13] using CRISPR/Cas9 systems. In rice, the natural PDS was supplemented by its bacterial homolog to create Golden Rice and thus increase the β-carotene content of the rice endosperm.
See also
- Phytoene desaturase (lycopene-forming)
- Phytoene desaturase (neurosporene-forming)
- Phytoene desaturase (zeta-carotene-forming)
- Phytoene desaturase (3,4-didehydrolycopene-forming)
References
- ↑ 1.0 1.1 PDB: 5mog; "Structure of Phytoene Desaturase Provides Insights into Herbicide Binding and Reaction Mechanisms Involved in Carotene Desaturation". Structure 25 (8): 1222–1232.e3. August 2017. doi:10.1016/j.str.2017.06.002. PMID 28669634.
- ↑ "Purification and reactivation of recombinant Synechococcus phytoene desaturase from an overexpressing strain of Escherichia coli". The Biochemical Journal 291 ( Pt 3) (3): 687–92. May 1993. doi:10.1042/bj2910687. PMID 8489496.
- ↑ "Phytoene desaturase: heterologous expression in an active state, purification, and biochemical properties". Protein Expression and Purification 10 (2): 175–9. July 1997. doi:10.1006/prep.1997.0730. PMID 9226712.
- ↑ "zeta-Carotene cis isomers as products and substrates in the plant poly-cis carotenoid biosynthetic pathway to lycopene". Planta 220 (5): 785–93. March 2005. doi:10.1007/s00425-004-1395-2. PMID 15503129.
- ↑ "Mechanistic aspects of carotenoid biosynthesis.". Chemical Reviews 114 (1): 164–193. 31 October 2013. doi:10.1021/cr400106y. PMID 24175570.
- ↑ "Phytoene Desaturase from Oryza sativa: Oligomeric Assembly, Membrane Association and Preliminary 3D-Analysis". PLOS ONE 10 (7): e0131717. July 2015. doi:10.1371/journal.pone.0131717. PMID 26147209. Bibcode: 2015PLoSO..1031717G.
- ↑ "Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation". The Plant Cell 7 (12): 2139–49. December 1995. doi:10.1105/tpc.7.12.2139. PMID 8718624.
- ↑ "Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis". Cell Research 17 (5): 471–82. May 2007. doi:10.1038/cr.2007.40. PMID 17486124.
- ↑ "Bleaching herbicide norflurazon inhibits phytoene desaturase by competition with the cofactors". Journal of Agricultural and Food Chemistry 49 (11): 5270–2. November 2001. doi:10.1021/jf0106751. PMID 11714315.
- ↑ Sandmann, Gerhard (2002). "2: Bleaching Herbicides: Action Mechanism in Carotenoid Biosynthesis, Structural Requirements and Engineering of Resistance". in Böger, Peter; Wakabayashi, Ko; Hirai, Kenji. Herbicide Classes in Development: Mode of Action, Targets, Genetic Engineering, Chemistry (1 ed.). Springer-Verlag Berlin Heidelberg. pp. 43–57. doi:10.1007/978-3-642-59416-8_2. ISBN 978-3-642-63972-2.
- ↑ "Efficient Genome Editing in Apple Using a CRISPR/Cas9 system". Scientific Reports 6: 31481. August 2016. doi:10.1038/srep31481. PMID 27530958. Bibcode: 2016NatSR...631481N.
- ↑ "CRISPR/Cas9-mediated targeted mutagenesis in grape". PLOS ONE 12 (5): e0177966. May 2017. doi:10.1371/journal.pone.0177966. PMID 28542349. Bibcode: 2017PLoSO..1277966N.
- ↑ Naim, Fatima; Dugdale, Benjamin; Kleidon, Jennifer; Brinin, Anthony; Shand, Kylie; Waterhouse, Peter; Dale, James (2018-07-09). "Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9". Transgenic Research 27 (5): 451–460. doi:10.1007/s11248-018-0083-0. ISSN 1573-9368. PMID 29987710.
External links
- 15-cis-phytoene+desaturase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |