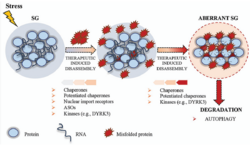
Biology:Stress granule
In cellular biology, stress granules are biomolecular condensates in the cytosol composed of proteins and RNAs that assemble into 0.1–2 μm membraneless organelles when the cell is under stress.[1][2][3] The mRNA molecules found in stress granules are stalled translation pre-initiation complexes associated with 40S ribosomal subunits, translation initiation factors, poly(A)+ mRNAs and RNA-binding proteins (RBPs). While they are membraneless organelles, stress granules have been proposed to be associated with the endoplasmatic reticulum.[4] There are also nuclear stress granules. This article is about the cytosolic variety.
Proposed functions
The function of stress granules remains largely unknown. Stress granules have long been proposed to have a function to protect RNAs from harmful conditions, thus their appearance under stress.[5] The accumulation of RNAs into dense globules could keep them from reacting with harmful chemicals and safeguard the information coded in their RNA sequence.
Stress granules might also function as a decision point for untranslated mRNAs. Molecules can go down one of three paths: further storage, degradation, or re-initiation of translation.[6] Conversely, it has also been argued that stress granules are not important sites for mRNA storage nor do they serve as an intermediate location for mRNAs in transit between a state of storage and a state of degradation.[7]
Efforts to identify all RNAs within stress granules (the stress granule transcriptome) in an unbiased way by sequencing RNA from biochemically purified stress granule "cores" have shown that RNAs are not recruited to stress granules in a sequence-specific manner, but rather generically, with longer and/or less-optimally translated transcripts being enriched.[8] These data imply that the stress granule transcriptome is influenced by the valency of RNA (for proteins or other RNAs) and by the rates of RNA run-off from polysomes. The latter is further supported by recent single molecule imaging studies.[9] Furthermore, it was estimated that only about 15% of the total mRNA in the cell is localized to stress granules,[8] suggesting that stress granules only influence a minority of mRNAs in the cell and may not be as important for mRNA processing as previously thought.[8][10] That said, these studies represent only a snapshot in time, and it is likely that a larger fraction of mRNAs are at one point stored in stress granules due to those RNAs transiting in and out.
The stress proteins that are the main component of stress granules in plant cells are molecular chaperones that sequester, protect, and possibly repair proteins that unfold during heat and other types of stress.[11][12] Therefore, any association of mRNAs with stress granules may simply be a side effect of the association of partially unfolded RNA-binding proteins with stress granules,[13] similar to the association of mRNAs with proteasomes.[14]
Formation
Environmental stressors trigger cellular signaling, eventually leading to the formation of stress granules. In vitro, these stressors can include heat, cold, oxidative stress (sodium arsenite), endoplasmic reticulum stress (thapsigargin), proteasome inhibition (MG132), hyperosmotic stress, ultraviolet radiation, inhibition of eIF4A (pateamine A, hippuristanol, or RocA), nitric oxide accumulation after treatment with 3-morpholinosydnonimine (SIN-1),[15] perturbation of pre-mRNA splicing,[16] and other stressors, like puromycin, which result in disassembled polysomes.[17] Many of these stressors result in the activation of particular stress-associated kinases (HRI, PERK, PKR, and GCN2), translational inhibition and stress granule formation.[17] Stress granules will also form upon Gαq activation in a mechanism that involves the release of stress granule associated proteins from the cytosolic population of the Gαq effector phospholipase Cβ.[18]
Stress granule formation is often downstream of the stress-activated phosphorylation of eukaryotic translation initiation factor eIF2α; this does not hold true for all types of stressors that induce stress granules,[17] for instance, eIF4A inhibition. Further downstream, prion-like aggregation of the protein TIA-1 promotes the formation of stress granules. The term prion-like is used because aggregation of TIA-1 is concentration dependent, inhibited by chaperones, and because the aggregates are resistant to proteases.[19] It has also been proposed that microtubules play a role in the formation of stress granules, perhaps by transporting granule components. This hypothesis is based on the fact that disruption of microtubules with the chemical nocodazole blocks the appearance of the granules.[20] Furthermore, many signaling molecules have been shown to regulate the formation or dynamics of stress granules; these include the "master energy sensor" AMP-activated protein kinase (AMPK),[21] the O-GlcNAc transferase enzyme (OGT),[22] and the pro-apoptotic kinase ROCK1.[23]
Potential roles of RNA-RNA interactions
RNA phase transitions driven in part by intermolecular RNA-RNA interactions may play a role in stress granule formation. Similar to intrinsically disordered proteins, total RNA extracts are capable of undergoing phase separation in physiological conditions in vitro.[24] RNA-seq analyses demonstrate that these assemblies share a largely overlapping transcriptome with stress granules,[24][8] with RNA enrichment in both being predominately based on the length of the RNA. Further, stress granules contain many RNA helicases,[25] including the DEAD/H-box helicases Ded1p/DDX3, eIF4A1, and RHAU.[26] In yeast, catalytic ded1 mutant alleles give rise to constitutive stress granules[27] ATPase-deficient DDX3X (the mammalian homolog of Ded1) mutant alleles are found in pediatric medulloblastoma,[28] and these coincide with constitutive granular assemblies in patient cells.[29] These mutant DDX3 proteins promote stress granule assembly in HeLa cells.[29] In mammalian cells, RHAU mutants lead to reduced stress granule dynamics.[26] Thus, some hypothesize that RNA aggregation facilitated by intermolecular RNA-RNA interactions plays a role in stress granule formation, and that this role may be regulated by RNA helicases.[30] There is also evidence that RNA within stress granules is more compacted, compared to RNA in the cytoplasm, and that the RNA is preferentially post-translationally modified by N6-methyladenosine (m6A) on its 5' ends.[31][32] Recent work has shown that the highly abundant translation initiation factor and DEAD-box protein eIF4A limits stress granule formation. It does so through its ability to bind ATP and RNA, acting analogously to protein chaperones like Hsp70.[33]
Connection with processing bodies
Stress granules and P-bodies (processing bodies) share RNA and protein components, both appear under stress, and can physically associate with one another. As of 2018, of the ~660 proteins identified as localizing to stress granules, ~11% also have been identified as processing body-localized proteins (see below). The protein G3BP1 is necessary for the proper docking of processing bodies and stress granules to each other, which may be important for the preservation of polyadenylated mRNAs.[34]
Although some protein components are shared between stress granules and processing bodies, the majority of proteins in either structure are uniquely localized to either structure.[35] While both stress granules and P-bodies are associated with mRNAs, processing bodies have been long proposed to be sites of mRNA degradation because they contain enzymes like DCP1/2 and XRN1 that are known to degrade mRNAs.[36] However, others have demonstrated that mRNAs associated with processing bodies are largely translationally repressed but not degraded.[35] It has also been proposed that mRNAs selected for degradation are passed from stress granules to processing bodies,[36] though there is also data suggesting that processing bodies precede and promote stress granule formation.[37]
Protein composition of stress granules
The complete proteome of stress granules is still unknown, but efforts have been made to catalog all of the proteins that have been experimentally demonstrated to transit into stress granules.[38][39][40] Importantly, different stressors can result in stress granules with different protein components.[17] Many stress granule-associated proteins have been identified by transiently stressing cultured cells and utilizing microscopy to detect the localization of a protein of interest either by expressing that protein fused to a fluorescent protein (i.e. green fluorescent protein (GFP)) and/or by fixing cells and using antibodies to detect the protein of interest along with known protein markers of stress granules (immunocytochemistry).[41]
In 2016, stress granule "cores" were experimentally identified and then biochemically purified for the first time. Proteins in the cores were identified in an unbiased manner using mass spectrometry. This technical advance lead to the identification of hundreds of new stress granule-localized proteins.[42][25][43]
The proteome of stress granules has also been experimentally determined by using two slightly different proximity labeling approaches. One of these proximity labeling approaches is the ascorbate peroxidase (APEX) method, in which cells are engineered to express a known stress granule protein, such as G3BP1, fused to a modified ascorbate peroxidase enzyme called APEX.[38][44] Upon incubating the cells in biotin and treating the cells with hydrogen peroxide, the APEX enzyme will be briefly activated to biotinylate all proteins in close proximity to the protein of interest, in this case G3BP1 within stress granules. Proteins that are biotinylated can then be isolated via streptavidin and identified using mass spectrometry. The APEX technique was used to identify ~260 stress granule-associated proteins in several cell types, including neurons, and with various stressors. Of the 260 proteins identified in this study, ~143 had not previously been demonstrated to be stress granule-associated.[44]
Another proximity labeling method used to determine the proteome of stress granules is BioID.[45] BioID is similar to the APEX approach, in that a biotinylating protein (BirA* instead of APEX) was expressed in cells as a fusion protein with several known stress granule-associated proteins. Proteins in close proximity to BirA* will be biotinylated and are then identified by mass spectrometry. Youn et al. used this method to identify/predict 138 proteins as stress granule-associated and 42 as processing body-associated.[45]
A curated database of stress granule-associated proteins can be found here [1].[40]
The following is a list of proteins that have been demonstrated to localize to stress granules (compiled from [38][39][25][44][45][46]):
| Gene ID | Protein Name | Description | References | Also found in processing bodies? |
|---|---|---|---|---|
| ABCF1 | ABCF1 | ATP Binding Cassette Subfamily F Member 1 | [44] | |
| ABRACL | ABRACL | ABRA C-Terminal Like | [44] | |
| ACAP1 | ACAP1 | ArfGAP With Coiled-Coil, Ankyrin Repeat And PH Domains 1 | [44] | |
| ACBD5 | ACBD5 | Acyl-CoA Binding Domain Containing 5 | [44] | |
| ACTBL2 | ACTBL2 | Beta-actin-like protein 2 | [25] | yes[35] |
| ACTR1A | ACTR1A | Alpha-centractin | [25] | |
| ACTR1B | ACTR1B | Beta-centractin | [25] | |
| ADAR | ADAR1 | Adenosine Deaminase, RNA Specific | [47][25] | |
| ADD1 | Adducin 1 | Adducin 1 | [44] | |
| AGO1 | Argonaute 1/EIF2C1 | Argonaute 1, RISC Catalytic Component | [44][48] | yes[35] |
| AGO2 | Argonaute 2 | Argonaute 2, RISC Catalytic Component | [44][49][48][50][25][51][46] | yes[35] |
| AKAP8 | AKAP8 | A-Kinase Anchoring Protein 8 | [46] | |
| AKAP9 | AKAP350 | A-Kinase Anchoring Protein 9 | [52] | |
| AKAP13 | AKAP13/LBC | A-Kinase Anchoring Protein 13 | [44][46] | |
| ALDH18A1 | ALDH18A1 | Delta-1-pyrroline-5-carboxylate synthase | [25] | |
| ALG13 | ALG13 | ALG13, UDP-N-Acetylglucosaminyltransferase Subunit | [45] | |
| ALPK2 | ALPK2/HAK | Alpha Kinase 2 | [46] | |
| AMOTL2 | AMOTL2/LCCP | Angiomotin Like 2 | [46] | |
| ANKHD1 | ANKHD1 | Ankyrin Repeat and KH Domain Containing 1 | [45] | yes[45] |
| ANKRD17 | ANKRD17/MASK2/GTAR | Ankyrin Repeat Domain 17 | [44][45] | yes[45] |
| ANG | Angiogenin | Angiogenin | [53] | |
| ANP32E | ANP32E | Acidic leucine-rich nuclear phosphoprotein 32 family member E | [25] | |
| ANXA1 | ANXA1 | Annexin A1 | [25] | |
| ANXA11 | ANXA11 | Annexin 11 | [44] | |
| ANXA6 | ANXA6 | Annexin 6 | [25] | |
| ANXA7 | ANXA7 | Annexin 7 | [25][44] | |
| APEX1 | APEX1 | DNA-(apurinic or apyrimidinic site) lyase | [25] | |
| APOBEC3C | APOBEC3C | Apolipoprotein B mRNA Editing Enzyme Catalytic Subunit 3C | [44][46] | |
| APOBEC3G | APOBEC3G | Apolipoprotein B mRNA Editing Enzyme Catalytic Subunit 3G | [48] | |
| ARID2 | ARID2/BAF200 | AT-Rich Interaction Domain 2 | [46] | |
| ARPC1B | ARPC1B | Actin-related protein 2/3 complex subunit 1B | [25] | |
| AHSA1 | AHA1 | Activator Of HSP90 ATPase Activity 1 | [54] | |
| AQR | AQR/IBP160 | Aquarius Intron-Binding Spliceosomal Factor | [44] | |
| ARMC6 | ARMC6 | Armadillo Repeat Containing 6 | [44] | |
| ASCC1 | ASCC1 | Activating Signal Cointegrator 1 Complex Subunit 1 | [44][45] | |
| ASCC3 | ASCC3 | Activating Signal Cointegrator 1 Complex Subunit 3 | [45] | |
| ATAD2 | ATAD2 | ATPase family AAA domain-containing protein 2 | [25] | |
| ATAD3A | ATAD3A | ATPase family AAA domain-containing protein 3A | [25] | yes[35] |
| ATG3 | ATG3 | Autophagy Related 3 | [44] | |
| ATP5A1 | ATP5A1 | ATP synthase subunit alpha, mitochondrial | [25] | |
| ATP6V1G1 | ATP6V1G1/ATP6G | ATPase H+ Transporting V1 Subunit G1 | [44] | |
| ATXN2 | Ataxin 2 | Ataxin 2 | ||
| ATXN2L | Ataxin-2 like | Ataxin 2 Like | [25][44][45][46][55][56] | |
| BAG3 | BAG3 | BAG family molecular chaperone regulator 3 | [25] | |
| BANF1 | BANF1 | Barrier-to-autointegration factor | [25] | |
| BAZ1B | BAZ1B | Bromodomain Adjacent To Zinc Finger Domain 1B | [46] | |
| BAZ2A | BAZ2A | Bromodomain Adjacent To Zinc Finger Domain 2A | [46] | |
| BCCIP | BCCIP | BRCA2 And CDKN1A Interacting Protein | [44] | |
| BCLAF1 | BCLAF1 | BCL2 Associated Transcription Factor 1 | [44] | |
| BICC1 | BICC1 | BicC Family RNA Binding Protein 1 | [45] | |
| BIRC2 | BIRC2/CIAP1 | Baculoviral IAP Repeat Containing 2 | [46] | |
| BLM | BLM | BLM RecQ Like Helicase | [46] | |
| BOD1L1 | BOD1L1/FAM44A | Biorientation Of Chromosomes In Cell Division 1 Like 1 | [46] | |
| BOLL | BOULE | Boule Homolog, RNA Binding Protein | [57] | |
| BRAT1 | BRAT1 | BRCA1-associated ATM activator 1 | [25] | |
| BRF1 | BRF1 | BRF1, RNA Polymerase III Transcription Initiation Factor Subunit | [36] | |
| BTG3 | BTG3 | BTG Anti-Proliferation Factor 3 | [45] | yes[45] |
| C9orf72 | C9orf72 | Uncharacterized protein C9orf72 | [58][59] | |
| C15orf52 | C15orf52 | Uncharacterized protein C15orf52 | [25] | |
| C20orf27 | C20orf72 | Chromosome 20 Open Reading Frame 27 | [44] | |
| C2CD3 | C2CD3 | C2 Calcium Dependent Domain Containing 3 | [44] | |
| CALML5 | CALML5 | Calmodulin-like protein 5 | [25] | |
| CALR | Calreticulin/CRT | Calreticulin | [60] | |
| CAMSAP1 | CAMSAP1 | Calmodulin Regulated Spectrin Associated Protein 1 | [46] | |
| CAP1 | CAP1 | Adenylyl cyclase-associated protein 1 | [25] | |
| CAPRIN1 | Caprin-1 | Cell Cycle Associated Protein 1 | [44][45][61][52][62][25][63][34][64][56][46] | |
| CAPZA2 | CAPZA2 | F-actin-capping protein subunit alpha-2 | [25] | |
| CAPZB | CAPZB | Capping Actin Protein Of Muscle Z-Line Subunit Beta | [46] | |
| CARHSP1 | CARHSP1 | Calcium-regulated heat stable protein 1 | [25] | |
| CASC3 | MLN51/BTZ | Cancer Susceptibility 3 | [44][45][46][65][66] | |
| CBFB | CBFB | Core-binding factor subunit beta | [25] | |
| CBS | CBS | Cystathionine Beta-Synthase | [46] | |
| CBX1 | CBX1 | Chromobox protein homolog 1 | [25][56] | |
| CBX3 | CBX3 | Chromobox protein homolog 3 | [46] | |
| CCAR1 | CARP-1 | Cell Division Cycle and Apoptosis Regulator 1 | [52][46] | |
| CCDC9 | CCDC9 | Coiled-Coil Domain Containing 9 | [46] | |
| CCDC9B | CCDC9B | Coiled-Coil Domain Containing 9B | [46] | |
| CCDC124 | CCDC124 | Coiled-Coil Domain Containing 124 | [44] | |
| CCDC85C | CCDC85C | Coiled-Coil Domain Containing 85C | [44] | |
| CCT3 | CCT3 | T-complex protein 1 subunit gamma | [25] | |
| CCT6A | CCT6A | T-complex protein 1 subunit zeta | [25] | |
| CDC20 | CDC20 | Cell Division Cycle 20 | [46] | |
| CDC37 | CDC37 | Cell Division Cycle 37 | [54] | |
| CDC5L | CDC5L | Cell division cycle 5-like protein | [25] | |
| CDC73 | CDC73 | Parafibromin | [25] | |
| CDK1 | CDK1 | Cyclin-dependent kinase 1 | [25] | |
| CDK2 | CDK2 | Cyclin Dependent Kinase 2 | [67] | |
| CDV3 | CDV3 | CDV3 Homolog | [44] | |
| CELF1 | CUGBP1 | CUGBP Elav-Like Family Member 1 | [25][44][45][46][68] | |
| CELF2 | CUGBP2/BRUNOL3 | CUGBP Elav-Like Family Member 2 | [44] | |
| CELF3 | CUGBP3/BRUNOL1 | CUGBP Elav-Like Family Member 3 | [44] | |
| CENPB | CENPB | Major centromere autoantigen B | [25] | |
| CENPF | CENPF | Centromere Protein F | [46] | |
| CEP78 | CEP78/CRDHL | Centrosomal Protein 78 | [44] | |
| CEP85 | CEP85/CCDC21 | Centrosomal Protein 78 | [45] | |
| CERKL | Ceramide-Kinase Like | Ceramide Kinase Like | [69] | |
| CFL1 | Cofilin-1 | Cofilin-1 | [25] | |
| CHCHD3 | CHCHD3 | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3, mitochondrial | [25] | |
| CHORDC1 | CHORDC1/CHP1 | Cysteine and histidine-rich domain-containing protein 1 | [25] | |
| CIRBP | CIRP | Cold Inducible RNA Binding Protein | [44][46][70] | |
| CIT | CIT | Citron Rho-interacting kinase | [25] | |
| CLIC4 | CLIC4 | Chloride intracellular channel protein 4 | [25] | |
| CLNS1A | CLNS1A | Chloride Nucleotide-Sensitive Channel 1A | [44] | |
| CLPP | CLPP | Caseinolytic Mitochondrial Matrix Peptidase Proteolytic Subunit | [44] | |
| CNBP | ZNF9 | CCHC-Type Zinc Finger Nucleic Acid Binding Protein | [46][71] | |
| CNN3 | CNN3 | Calponin-3 | [25] | |
| CNOT1 | CNOT1/CCR4 | CCR4-Not Transcription Complex Subunit 1 | [25][45] | yes[45][72] |
| CNOT10 | CNOT10 | CCR4-Not Transcription Complex Subunit 10 | [45] | yes[45] |
| CNOT11 | CNOT11 | CCR4-Not Transcription Complex Subunit 11 | [45] | yes[45] |
| CNOT2 | CNOT2 | CCR4-Not Transcription Complex Subunit 2 | [45] | yes[45] |
| CNOT3 | CNOT3 | CCR4-Not Transcription Complex Subunit 3 | [45] | yes[45] |
| CNOT4 | CNOT4 | CCR4-Not Transcription Complex Subunit 4 | [45] | yes[45] |
| CNOT6 | CNOT6 | CCR4-Not Transcription Complex Subunit 6 | [45] | yes[45] |
| CNOT6L | CNOT6L | CCR4-Not Transcription Complex Subunit 6L | [45] | yes[45] |
| CNOT7 | CNOT7 | CCR4-Not Transcription Complex Subunit 7 | [45] | yes[45] |
| CNOT8 | CNOT8 | CCR4-Not Transcription Complex Subunit 8 | [45] | yes[45] |
| CNOT9 | CNOT9 | CCR4-Not Transcription Complex Subunit 9 | [45] | |
| CORO1B | CORO1B | Coronin-1B | [25] | |
| CPB2 | Carboxypeptidase B2 | Carboxypeptidase B2 | [73] | |
| CPEB1 | CPEB | Cytoplasmic Polyadenylation Element Binding Protein 1 | ||
| CPEB4 | CPEB4 | Cytoplasmic Polyadenylation Element Binding Protein 4 | [44][45][46] | yes[45] |
| CPSF3 | CPSF3 | Cleavage and polyadenylation specificity factor subunit 3 | [25] | |
| CPSF6 | CPSF6 | Cleavage and polyadenylation specificity factor subunit 6 | [25] | |
| CPSF7 | CPSF7 | Cleavage and polyadenylation specificity factor subunit 7 | [25] | |
| CPVL | CPVL | Carboxypeptidase, Vitellogenic Like | [45] | yes[45] |
| CRKL | CRKL | CRK Like Proto-Oncogene, Adaptor Protein | [44] | |
| CROCC | CROCC | Ciliary Rootlet Coiled-Coil, Rootletin | [44] | |
| CRYAB | CRYAB | Alpha-crystallin B chain | [25] | |
| CRYBG1 | CRYBG1 | Crystallin Beta-Gamma Domain Containing 1 | [46] | |
| CSDE1 | CSDE1 | Cold shock domain-containing protein E1 | [25][44][45][46][56] | |
| CSE1L | CSE1L/XPO2/Exportin-2 | Exportin-2 | [25] | |
| CSNK2A1 | Casein Kinase 2 alpha | Casein Kinase 2 Alpha 1 | [74] | |
| CSTB | Cystatin B | Cystatin B | [44] | |
| CSTF1 | CSTF1 | Cleavage stimulation factor subunit 1 | [25] | |
| CTNNA2 | CTNNA2 | Catenin alpha-2 | [25] | |
| CTNND1 | CTNND1 | Catenin delta-1 | [25] | |
| CTTNBP2NL | CTTNBP2NL | CTTNBP2 N-terminal-like protein | [25] | |
| CWC22 | CWC22 | Pre-mRNA-splicing factor CWC22 homolog | [25] | |
| DAZAP1 | DAZAP1 | DAZ-associated protein 1 | [25][44][45][46] | |
| DAZAP2 | PRTB | DAZ Associated Protein 2 | [75] | |
| DAZL | DAZL1 | Deleted In Azoospermia Like | [76] | |
| DCD | DCD | Dermcidin | [25] | |
| DCP1A | DCP1a | Decapping mRNA 1a | [25][44][77] | yes[35] |
| DCP1B | DCP1b | Decapping mRNA 1b | [44][46] | yes[35] |
| DCP2 | DCP2 | Decapping mRNA 2 | [45] | |
| DCTN1 | DCTN1 | Dynactin subunit 1 | [25] | |
| DDX1 | DEAD box protein 1 | DEAD-Box Helicase 1 | [25][44][45][46][78] | |
| DDX11 | DEAD box protein 11 | DEAD-Box Helicase 11 | [46] | |
| DDX19A | DDX19A | ATP-dependent RNA helicase DDX19A | [25][56] | |
| DDX21 | DDX21 | Nucleolar RNA helicase 2 | [25] | yes[35] |
| DDX3 | DEAD box protein 3 | DEAD-Box Helicase 3 | [25][79][80] | |
| DDX3X | DDX3X | DEAD-Box Helicase 3, X-Linked | [44][45][46][81][82][56] | |
| DDX3Y | DDX3Y | DEAD-Box Helicase 3, Y-Linked | [44] | |
| DDX31 | DDX31 | DEAD-Box Helicase 31 | [46] | |
| DDX47 | DDX47 | Probable ATP-dependent RNA helicase DDX47 | [25] | |
| DDX50 | DDX50 | ATP-dependent RNA helicase DDX50 | [25] | yes[35] |
| DDX58 | RIG-I | DExD/H-Box Helicase 58 | [83] | |
| DDX6 | DEAD box protein 6 | DEAD-Box Helicase 6 | [25][44][45][84][85][77][48][86][46] | yes[35][45] |
| DERA | DERA | Deoxyribose-Phosphate Aldolase | [87] | |
| DGCR8 | DGCR8 | DGCR8 Microprocessor Complex Subunit | [46] | |
| DHX30 | DHX30 | Putative ATP-dependent RNA helicase DHX30 | [25][44] | yes[35] |
| DHX33 | DHX33 | DEAH-Box Helicase 33 | [44] | |
| DHX36 | RHAU | DEAH-Box Helicase 36 | [44][45][26][46] | |
| DHX57 | DHX57 | DExH-Box Helicase 57 | [45][46] | |
| DHX58 | LGP2 | DExH-Box Helicase 58 | [83] | |
| DIDO1 | DIDO1 | Death Inducer-Obliterator 1 | [46] | |
| DIS3L2 | DIS3L2/FAM3A | DIS3 Like 3'-5' Exoribonuclease 2 | [44] | |
| DISC1 | Disrupted in Schizophrenia 1 | Disrupted In Schizophrenia 1 | ||
| DKC1 | DKC1 | dyskerin; H/ACA ribonucleoprotein complex subunit 4 | ||
| DNAI1 | Axonemal Dynein Intermediate Chain 1 | Dynein Axonemal Intermediate Chain 1 | [88] | |
| DNAJA1 | DNAJA1 | DnaJ homolog subfamily A member 1 | [25] | |
| DNAJC8 | DNAJC8 | DnaJ homolog subfamily C member 8 | [25] | |
| DOCK4 | DOCK4 | Dedicator Of Cytokinesis 4 | [46] | |
| DPYSL2 | DPYSL2 | Dihydropyrimidinase-related protein 2 | [25] | |
| DPYSL3 | DPYSL3 | Dihydropyrimidinase-related protein 3 | [25] | |
| DROSHA | DROSHA | Drosha Ribonuclease III | [44] | |
| DSP | DSP | Desmoplakin | [25][44] | |
| DST | DST | Dystonin | [25] | |
| DSTN | DSTN | Destrin | [25] | |
| DTL | DTL | Denticleless E3 Ubiquitin Protein Ligase Homolog | [46] | |
| DTX3L | DTX3L | E3 ubiquitin-protein ligase DTX3L | [25] | |
| DUSP12 | DUSP12/YVH1 | Dual Specificity Phosphatase 12 | [89] | |
| DYNC1H1 | Cytoplasmic Dynein Heavy Chain 1 | Dynein Cytoplasmic 1 Heavy Chain 1 | [88] | |
| DYNLL1 | Cytoplasmic Dynein Light Polypeptide | Dynein Light Chain LC8-Type 1 | [44][90] | |
| DYNLL2 | DYNLL2 | Dynein light chain 2, cytoplasmic | [25] | |
| DYRK3 | DYRK3 | Dual Specificity Tyrosine Phosphorylation Regulated Kinase 3 | [91] | |
| DZIP1 | DZIP1 | DAZ Interacting Zinc Finger Protein 1 | [92] | |
| DZIP3 | DZIP3 | DAZ Interacting Zinc Finger Protein 3 | [45] | |
| EDC3 | EDC3 | Enhancer of mRNA Decapping 3 | [44][45][46] | yes[45] |
| EDC4 | EDC4 | Enhancer of mRNA-Decapping protein 4 | [25][44][46] | yes[35] |
| EIF1 | EIF1 | Eukaryotic Translation Initiation Factor 1 | [44] | |
| EIF2A | EIF2A | Eukaryotic Translation Initiation Factor 2A | [36][25][52][93] | |
| EIF2AK2 | Protein Kinase R/PKR | Eukaryotic Translation Initiation Factor 2 Alpha Kinase 2 | [64][83][94] | |
| EIF2B1-5 | EIF2B | Eukaryotic Translation Initiation Factor 2B | [93] | |
| EIF2S1 | EIF2A subunit 1 | Eukaryotic Translation Initiation Factor 2 Subunit Alpha | [25] | |
| EIF2S2 | EIF2A subunit 2 | Eukaryotic Translation Initiation Factor 2 Subunit Beta | [25] | |
| EIF3A | EIF3A | Eukaryotic Translation Initiation Factor 3 Subunit A | [25][44][49][34][95][46] | |
| EIF3B | EIF3B | Eukaryotic Translation Initiation Factor 3 Subunit B | [36][25][75][96][97] | |
| EIF3C | EIF3C | Eukaryotic Translation Initiation Factor 3 Subunit C | [44] | |
| EIF3D | EIF3D | Eukaryotic translation initiation factor 3 subunit D | [25][44][56] | |
| EIF3E | EIF3E | Eukaryotic translation initiation factor 3 subunit E | [25][44][56] | |
| EIF3F | EIF3F | Eukaryotic translation initiation factor 3 subunit F | [25] | |
| EIF3G | EIF3G | Eukaryotic translation initiation factor 3 subunit G | [25][44][56][46] | |
| EIF3H | EIF3H | Eukaryotic translation initiation factor 3 subunit H | [25][44][46] | |
| EIF3I | EIF3I | Eukaryotic translation initiation factor 3 subunit I | [25][46] | |
| EIF3J | EIF3J | Eukaryotic translation initiation factor 3 subunit J | [25][44] | |
| EIF3K | EIF3K | Eukaryotic translation initiation factor 3 subunit K | [25] | |
| EIF3L | EIF3L | Eukaryotic translation initiation factor 3 subunit L | [25][44][56] | |
| EIF3M | EIF3M | Eukaryotic translation initiation factor 3 subunit M | [25] | |
| EIF4A1 | EIF4A1 | Eukaryotic Translation Initiation Factor 4A1 | [25][44][98][46] | |
| EIF4A2 | EIF4A2 | Eukaryotic Translation Initiation Factor 4A2 | [44][99][46] | |
| EIF4A3 | EIF4A3 | Eukaryotic Translation Initiation Factor 4A3 | [44] | |
| EIF4B | EIF4B | Eukaryotic translation Initiation factor 4B | [25][44][46] | |
| EIF4E | EIF4E | Eukaryotic Translation Initiation Factor 4E | [95][93][4][100][66][101][102][36] | yes[36] |
| EIF4E2 | EIF4E2 | Eukaryotic Translation Initiation Factor 4E Family Member 2 | [45][102] | yes[45] |
| EIF4E3 | EIF4E3 | Eukaryotic Translation Initiation Factor 4E Family Member 3 | [102] | |
| EIF4ENIF1 | EIF4ENIF1 | Eukaryotic Translation Initiation Factor 4E Nuclear Import Factor 1 | [44][45] | yes[45] |
| EIF4G1 | EIF4G1 | Eukaryotic Translation Initiation Factor 4G1 | [25][44][95][93][4][100][103][104][75][105][34][46] | |
| EIF4G2 | EIF4G2 | Eukaryotic Translation Initiation Factor 4G2 | [25][45] | |
| EIF4G3 | EIF4G3 | Eukaryotic Translation Initiation Factor 4G3 | [44] | |
| EIF4H | EIF4H | Eukaryotic translation Initiation factor 4H | [25][44][46] | |
| EIF5A | EIF5A | Eukaryotic Translation Initiation Factor 5A | [96] | |
| ELAVL1 | HuR | ELAV Like RNA Binding Protein 1 | [25][34][44][106][95][107][100][101][75][90][108][109][46] | yes[35] |
| ELAVL2 | ELAVL2 | ELAV-like protein 2 | [25][44] | yes[35] |
| ELAVL3 | ELAVL3/HuC | ELAV Like RNA Binding Protein 3 | [44] | |
| ELAVL4 | HuD | ELAV Like RNA Binding Protein 4 | [44][110] | |
| ENC1 | ENC1 | Ectodermal-Neural Cortex 1 | [46] | |
| ENDOV | EndoV | Endonuclease V | [111] | |
| ENTPD1 | ENTPD1 | Ectonucleoside Triphosphate Diphosphohydrolase 1 | [44] | |
| EP400 | EP400 | E1A Binding Protein P400 | [46] | |
| EPPK1 | EPPK1 | Epiplakin | [25] | |
| ETF1 | ETF1 | Eukaryotic peptide chain release factor subunit 1 | [25] | |
| EWSR1 | EWSR1 | EWS RNA Binding Protein 1 | [112][113][46] | |
| FABP5 | FABP5 | Fatty Acid Binding Protein 5 | [44] | |
| FAM120A | FAM120A/OSSA | Constitutive coactivator of PPAR-gamma-like protein 1 | [25][44][45] | yes[35] |
| FAM120C | FAM120C | Family With Sequence Similarity 120C | [44][45] | |
| FAM168A | FAM168A | Family With Sequence Similarity 168 Member A | [46] | |
| FAM168B | FAM168B/MANI | Family With Sequence Similarity 168 Member B | [44] | |
| FAM83H | FAM83H | Family With Sequence Similarity 83 Member H | [46] | |
| FAM98A | FAM98A | Family With Sequence Similarity 98 Member A | [25][44][114][46] | |
| FAM98C | FAM98C | Family With Sequence Similarity 98 Member C | [46] | |
| FASTK | FAST | Fas Activated Serine/Threonine Kinase | [36] | yes[36] |
| FBL | FBL | rRNA 2-O-methyltransferase fibrillarin | [25] | |
| FBRSL1 | Fibrosin Like 1 | Fibrosin Like 1 | [45] | |
| FHL1 | FHL1 | Four and a half LIM domains protein 1 | [25] | |
| FKBP1A | FKBP1A | FKBP Prolyl Isomerase 1A | [46] | |
| FLNB | FLNB | Filamin-B | [25] | |
| FMR1 | FMRP | Fragile X Mental Retardation 1 | [23][25][44][45][65][66][100][115][116][89][56][46] | |
| FNDC3B | FNDC3B | Fibronectin type III domain-containing protein 3B | [25][45][46] | |
| FSCN1 | FSCN1 | Fascin | [25] | |
| FTSJ3 | FTSJ3 | pre-rRNA processing protein FTSJ3 | [25] | |
| FUBP1 | FUBP1 | Far Upstream Element Binding Protein 1 | [44][46] | |
| FUBP3 | FUBP3 | Far upstream element-binding protein 3 | [25][44][45][46] | |
| FUS | FUS | FUS RNA Binding Protein | ||
| FXR1 | FXR1 | FMR1 Autosomal Homolog 1 | [25][44][45][115][100][101][117][46] | |
| FXR2 | FXR2 | FMR1 Autosomal Homolog 2 | [25][44][45][115][100][46] | |
| G3BP1 | G3BP1 | G3BP Stress Granule Assembly Factor 1 | ||
| G3BP2 | G3BP2 | G3BP Stress Granule Assembly Factor 2 | [25][44][45][118][119][56][46] | |
| GABARAPL2 | GABARAPL2/GEF2/ATG8 | GABA Type A Receptor Associated Protein Like 2 | [44] | |
| GAK | GAK | Cyclin G Associated Kinase | [46] | |
| GAR1 | GAR1 | H/ACA Ribonucleoprotein Complex Subunit 1 | [120] | |
| GCA | Grancalcin | Grancalcin | [44] | |
| GEMIN5 | Gemin-5 | Gem Nuclear Organelle Associated Protein 5 | [103] | |
| GFPT1 | GFPT1 | Glutamine—fructose-6-phosphate aminotransferase [isomerizing] 1 | [25] | |
| GIGYF1 | GIGYF1/PERQ1 | GRB10 Interacting GYF Protein 1 | [44] | |
| GIGYF2 | GIGYF2/TNRC15/PARK11/PERQ2 | GRB10 Interacting GYF Protein 2 | [44][45] | yes[45] |
| GLE1 | GLE1 | GLE1, RNA Export Mediator | [45][121][122] | |
| GLO1 | Glyoxalase | Glyoxalase | [44] | |
| GLRX3 | GLRX3/Glutaredoxin 3/TNLX2 | Glutaredoxin 3 | [44] | |
| GLUD1 | GLUD1 | Glutamate Dehydrogenase 1 | [46] | |
| GNB2 | GNB2 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-2 | [25] | |
| GOLGA2 | Golgin A2 | Golgin A2 | [44] | |
| GPAT3 | GPAT3 | Glycerol-3-Phosphate Acyltransferase 3 | [46] | |
| GRB2 | GRB2/ASH | Growth Factor Receptor Bound Protein 2 | [44] | |
| GRB7 | GRB7 | Growth Factor Receptor Bound Protein 7 | [123][124] | |
| GRSF1 | GRSF1 | G-Rich RNA Sequence Binding Factor 1 | [44][45] | |
| GSPT1 | eRF3 | G1 To S Phase Transition 1 | [44][125] | |
| GTF2I | GTF2I | General Transcription Factor IIi | [46] | |
| GTF3C1 | GTF3C1 | General Transcription Factor IIIC Subunit 1 | [46] | |
| GTF3C4 | GTF3C4 | General Transcription Factor IIIC Subunit 4 | [46] | |
| H1F0 | H1F0 | Histone H1.0 | [25] | |
| H1FX | H1FX | Histone H1x | [25] | |
| H2AFV | H2AFV | Histone H2A.V | [25] | |
| HABP4 | Ki-1/57 | Hyaluronan Binding Protein 4 | [126] | |
| HDAC6 | HDAC6 | Histone Deacetylase 6 | [82][127][56] | |
| HDLBP | HDL-Binding Protein/VGL/Vigilin | High Density Lipoprotein Binding Protein | [44] | |
| HELZ | HELZ | Probable helicase with zinc finger domain | [25][44][45] | yes[45] |
| HELZ2 | HELZ2 | Helicase with zinc finger domain 2 | [25] | |
| HMGA1 | HMGA1 | High mobility group protein HMG-I/HMG-Y | [25] | |
| HMGB3 | HMGB3 | High mobility group protein B3 | [25] | |
| HMGN1 | HMGN1 | Non-histone chromosomal protein HMG-14 | [25] | |
| HNRNPA1 | HnRNPA1 | Heterogeneous Nuclear Ribonucleoprotein A1 | [25][44][49][128][129][130][131] | |
| HNRNPA2B1 | HnRNPA2/B1 | Heterogeneous Nuclear Ribonucleoprotein A2/B1 | [25][44][132][56] | |
| HNRNPA3 | HNRNPA3 | Heterogeneous nuclear ribonucleoprotein A3 | [25][44] | |
| HNRNPAB | HNRNPAB | Heterogeneous nuclear ribonucleoprotein A/B | [25][44][45] | |
| HNRNPD | HNRNPD | Heterogeneous nuclear ribonucleoprotein D | [44] | |
| HNRNPDL | HNRNPDL | Heterogeneous nuclear ribonucleoprotein D-like | [44] | |
| HNRNPF | HNRNPF | Heterogeneous nuclear ribonucleoprotein F | [44] | |
| HNRNPH1 | HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H1 | [44] | |
| HNRNPH2 | HNRNPH2 | Heterogeneous nuclear ribonucleoprotein H2 | [25] | |
| HNRNPH3 | HNRNPH3 | Heterogeneous nuclear ribonucleoprotein H3 | [44] | |
| HNRNPK | HNRNPK | Heterogeneous Nuclear Ribonucleoprotein K | [25][109][133] | |
| HNRNPUL1 | HNRNPUL1 | Heterogeneous nuclear ribonucleoprotein U-like protein 2 | [25] | |
| HSBP1 | HSBP1 | Heat Shock Factor Binding Protein 1 | [44] | |
| HSP90AA1 | HSP90 | Heat shock protein HSP 90-alpha | [25] | |
| HSPA4 | HSP70 RY | Heat shock 70 kDa protein 4 | [25] | |
| HSPA9 | HSP70 9B | Stress-70 protein, mitochondrial | [25] | |
| HSPB1 | HSP27 | Heat Shock Protein Family B (Small) Member 1 | yes[35] | |
| HSPB8 | HSPB8 | Heat Shock Protein Family B (Small) Member 8 | [134] | |
| HSPBP1 | HSPBP1 | HSPA (Hsp70) Binding Protein 1 | [135] | |
| HSPD1 | HSPD1 | 60 kDa heat shock protein, mitochondrial | [25][44] | |
| HTT | Huntingtin | Huntingtin | [62] | |
| IBTK | IBTK | Inhibitor Of Bruton Tyrosine Kinase | [45] | |
| IFIH1 | MDA5 | Interferon Induced With Helicase C Domain 1 | [83] | |
| IGF2BP1 | IGF2BP1 | Insulin-like Growth Factor 2 mRNA-binding protein 1 | [25][44][45] | yes[35] |
| IGF2BP2 | IGF2BP2 | Insulin-like Growth Factor 2 mRNA-binding protein 2 | [25][44][45] | yes[35] |
| IGF2BP3 | IGF2BP3 | Insulin-like Growth Factor 2 mRNA Binding Protein 3 | [25][44][45][118] | yes[35] |
| IK | IK | Protein Red | [25] | |
| ILF3 | NF90 | Interleukin Enhancer Binding Factor 3 | [136] | yes[35] |
| IPO7 | IPO7 | Importin-7 | [25] | |
| IPPK | IP5K | Inositol-Pentakisphosphate 2-Kinase | [137] | |
| ITGB1 | ITGB1 | Integrin beta-1 | [25] | |
| JMJD6 | JMJD6 | Arginine Demethylase and Lysine Hydroxylase | [138] | |
| KANK2 | KANK2 | KN motif and ankyrin repeat domain-containing protein 2 | [25] | |
| KEAP1 | KEAP1/KLHL19 | Kelch Like ECH Associated Protein 1 | [44] | |
| KHDRBS1 | Sam68 | KH RNA Binding Domain Containing, Signal Transduction Associated 1 | [25][139][140][141] | |
| KHDRBS3 | KHDRBS3 | KH domain-containing, RNA-binding, signal transduction-associated protein 3 | [25] | |
| KHSRP | KSRP/FBP2 | KH-Type Splicing Regulatory Protein | [25][44][142] | |
| KIAA0232 | KIAA0232 | KIAA0232 | [45] | yes[45] |
| KIAA1524 | CIP2A | Protein CIP2A | [25] | |
| KIF1B | KIF1B | Kinesin Family Member 1B | [45] | |
| KIF13B | KIF13B/GAKIN | Kinesin Family Member 13B | [44] | |
| KIF23 | KIF23 | Kinesin-like protein KIF23 | [25] | yes[35] |
| KIF2A | Kinesin Heavy Chain Member 2 | Kinesin Family Member 2A | [88] | |
| KLC1 | Kinesin Light Chain 1 | Kinesin Light Chain 1 | [88] | |
| KPNA1 | Importin-ɑ5 | Karyopherin Subunit Alpha 1 | [25][44][143] | |
| KPNA2 | Importin-ɑ1 | Karyopherin Subunit Alpha 2 | [25][143][144][122] | |
| KPNA3 | Importin-ɑ4 | Karyopherin Subunit Alpha 3 | [44][143] | |
| KPNA6 | Importin-ɑ7 | Importin subunit alpha | [25] | |
| KPNB1 | Importin-β1 | Karyopherin Subunit Beta 1 | [25][143][122][56] | |
| L1RE1 | LINE1 ORF1p | LINE1 ORF1 protein | [25][49] | |
| LANCL1 | LanC Like 1 | LanC Like 1 | [44] | |
| LARP1 | LARP1 | La-related protein 1 | [25] | |
| LARP1B | LARP1B | La-related protein 1b | [45] | |
| LARP4 | La-Related protein 4 | La Ribonucleoprotein Domain Family Member 4 | [25][44][45][145] | |
| LARP4B | LARP4B | La Ribonucleoprotein Domain Family Member 4B | [44][45] | |
| LASP1 | LIM And SH3 Protein 1/MLN50 | LIM And SH3 Protein 1 | [44] | |
| LBR | LBR | Lamin-B receptor | [25] | |
| LEMD3 | LEMD3 | Inner nuclear membrane protein Man1 | [25] | |
| LIG3 | DNA Ligase 3 | DNA Ligase 3 | [44] | |
| LIN28A | LIN28A | Lin-28 Homolog A | [44][146] | |
| LIN28B | LIN28B | Lin-28 Homolog B | [44][146] | |
| LMNA | LMNA | Prelamin-A/C | [25] | |
| LPP | LPP | Lipoma-preferred partner | [25] | |
| LSM1 | LSM1 | LSM1 Homolog, mRNA Degradation Associated | [44] | yes[147] |
| LSM12 | LSM12 | LSM12 Homolog | [44][45] | |
| LSM14A | RAP55 | LSM14A, mRNA Processing Body Assembly Factor | [25][44][45][148][149] | yes[35][45] |
| LSM14B | LSM14B | Protein LSM14 homolog B | [25][44][45] | yes[35] |
| LSM3 | LSM3 | U6 snRNA-associated Sm-like protein LSm3 | [25] | yes[147] |
| LUC7L | LUC7L | Putative RNA-binding protein Luc7-like 1 | [25] | |
| LUZP1 | LUZP1 | Leucine zipper protein 1 | [25][45] | |
| MACF1 | MACF1 | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 | [25][56] | |
| MAEL | MAEL | Maelstrom Spermatogenic Transposon Silencer | [150] | |
| MAGEA4 | MAGEA4 | Melanoma-associated antigen 4 | [25] | |
| MAGED1 | MAGED1 | Melanoma-associated antigen D1 | [25][44][45] | |
| MAGED2 | MAGED2 | Melanoma-associated antigen D2 | [25] | |
| MAGOHB | MAGOHB | Protein mago nashi homolog 2 | [25] | |
| MAP1LC3A | LC3-I | Microtubule Associated Protein 1 Light Chain 3 Alpha | ||
| MAP4 | MAP4 | Microtubule-associated protein 4 | [25] | |
| MAPK1IP1L | MAPK1IP1L | Mitogen-Activated Protein Kinase 1 Interacting Protein 1 Like | [44] | |
| MAP4K4 | MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | [25] | |
| MAPK8 | JNK1 | Mitogen-Activated Protein Kinase 8 | [151] | |
| MAPRE1 | MAPRE1 | Microtubule-associated protein RP/EB family member 1 | [25] | |
| MAPRE2 | MAPRE2 | Microtubule Associated Protein RP/EB Family Member 2 | [44] | |
| MARF1 | MARF1 | Meiosis Regulator And mRNA Stability Factor 1 | [45] | yes[45] |
| MARS | MARS | Methionine—tRNA ligase, cytoplasmic | [25] | |
| MBNL1 | MBNL1 | Muscleblind Like Splicing Regulator 1 | [78] | |
| MBNL2 | MBNL2 | Muscleblind Like Splicing Regulator 2 | [45] | |
| MCM4 | MCM4 | DNA replication licensing factor MCM4 | [25] | |
| MCM5 | MCM5 | DNA replication licensing factor MCM5 | [25] | |
| MCM7 | MCM7 | DNA replication licensing factor MCM7 | [25] | yes[35] |
| METAP1 | METAP1 | Methionine aminopeptidase | [25] | |
| METAP2 | METAP2 | Methionyl Aminopeptidase 2 | [44] | |
| MCRIP1 | FAM195B/GRAN2 | Granulin-2 | [44][45][86] | |
| MCRIP2 | FAM195A/GRAN1 | Granulin-1 | [45][86] | |
| MEX3A | MEX3A | RNA-binding protein MEX3A | [25] | yes[35] |
| MEX3B | MEX3B | Mex-3 RNA Binding Family Member B | [44][152] | |
| MEX3C | MEX3C | Mex-3 RNA Binding Family Member C | [44][153] | |
| MEX3D | MEX3D | Mex-3 RNA Binding Family Member D | [45] | |
| MFAP1 | MFAP1 | Microfibrillar-associated protein 1 | [25] | |
| MKI67 | MKI67 | Antigen KI-67 | [25] | |
| MKRN2 | MKRN2 | Makorin Ring Finger Protein 2 | [44][45] | |
| MOV10 | MOV-10 | Mov10 RISC Complex RNA Helicase | [25][45][48] | yes[35][45] |
| MSH6 | MSH6 | DNA mismatch repair protein Msh6 | [25] | |
| MSI1 | Musashi-1 | Musashi RNA Binding Protein 1 | [44][149][154] | yes[35] |
| MSI2 | MSI2 | RNA-binding protein Musashi homolog 2 | [25][44] | |
| MTHFD1 | MTHFD1 | C-1-tetrahydrofolate synthase, cytoplasmic | [25] | |
| MTHFSD | MTHFSD | Methenyltetrahydrofolate Synthetase Domain Containing | [155] | |
| MTOR | MTOR | Mechanistic Target Of Rapamycin | [91][156] | |
| MYO6 | MYO6 | Unconventional myosin-VI | [25] | |
| NCOA3 | SRC-3 | Nuclear Receptor Coactivator 3 | [157] | |
| NDEL1 | NUDEL/MITAP1/EOPA | NudE Neurodevelopment Protein 1 Like 1 | [44] | |
| NELFE | NELF-E/RD | Negative Elongation Factor Complex Member E | [44] | |
| NEXN | NEXN | Nexilin | [25] | |
| NXF1 | NXF1/MEX67/TAP | Nuclear RNA Export Factor 1 | [45][56] | |
| NKRF | NRF | NFK-B Repressing Factor | [44] | |
| NOLC1 | Nucleolar And Coiled-Body Phosphoprotein 1/NOPP140 | Nucleolar And Coiled-Body Phosphoprotein 1 | [44] | |
| NONO | NonO | Non-POU Domain Containing Octamer Binding | [25][158] | |
| NOP58 | NOP58 | Nucleolar protein 58 | [25] | yes[35] |
| NOSIP | NOSIP | Nitric oxide synthase-interacting protein | [25] | |
| NOVA2 | NOVA2 | NOVA Alternative Splicing Regulator 2 | [44] | |
| NRG2 | Neuregulin-2 | Neuregulin-2 | [97] | |
| NSUN2 | NSUN2 | tRNA (cytosine(34)-C(5))-methyltransferase | [25] | |
| NTMT1 | NTMT1 | N-terminal Xaa-Pro-Lys N-methyltransferase 1 | [25] | |
| NUDC | NUDC | Nuclear migration protein nudC | [25] | |
| NUFIP1 | NUFIP | NUFIP1, FMR1 Interacting Protein 1 | [100] | |
| NUFIP2 | NUFIP2 | Nuclear fragile X mental retardation-interacting protein 2 | [25][44][45][86][56] | |
| NUPL2 | NUPL2 | Nucleoporin Like 2 | [122] | |
| NUP153 | NUP153 | Nucleoporin 153 | [44] | |
| NUP205 | NUP205 | Nuclear pore complex protein Nup205 | [25][122] | |
| NUP210 | NUP210/GP210 | Nucleoporin 210 | [122] | |
| NUP214 | NUP214 | Nucleoporin 214 | [122] | |
| NUP50 | NUP50 | Nucleoporin 50 | [122] | |
| NUP58 | NUP58/NUPL1 | Nucleoporin 58 | [122] | |
| NUP85 | NUP85 | Nucleoporin 85 | [122] | |
| NUP88 | NUP88 | Nucleoporin 88 | [122] | |
| NUP98 | NUP98/NUP96 | Nuclear pore complex protein Nup98-Nup96 | [25][122][56] | |
| OASL | OASL/OASL1 | 2'-5'-Oligoadenylate Synthetase Like | [159] | |
| OAS1 | OAS | 2′–5′ oligoadenylate synthetase | [83] | |
| OAS2 | OAS2 | 2'-5'-Oligoadenylate Synthetase 2 | [94] | |
| OGFOD1 | TPA1 | 2-Oxoglutarate And Iron Dependent Oxygenase Domain Containing 1 | [160] | |
| OGG1 | OGG1 | 8-Oxoguanine DNA Glycosylase | [161] | |
| OSBPL9 | Oxysterol Binding Protein Like 9 | Oxysterol Binding Protein Like 9 | [44] | |
| OTUD4 | OTUD4/HIN1 | OTU Deubiquitinase 4 | [44][45][162] | |
| P4HB | Prolyl 4-Hydroxylase Subunit Beta | Prolyl 4-Hydroxylase Subunit Beta | [44] | |
| PABPC1 | PABP1 | Poly(A) Binding Protein Cytoplasmic 1 | [25][44][45][163][107][164][115][66][100][118] | |
| PABPC4 | PABPC4 | Polyadenylate-binding protein 4 | [25][44][45] | |
| PAK4 | PAK4 | Serine/threonine-protein kinase PAK 4 | [25][44] | |
| PALLD | Palladin | Palladin | [25] | |
| PARG | PARG/PARG99/PARG102 | Poly(ADP-Ribose) Glycohydrolase | [165] | |
| PARK7 | PARK7/DJ-1 | Parkinsonism Associated Deglycase | [166] | yes[166] |
| PARN | PARN/DAN | Poly(A)-Specific Ribonuclease | [44] | |
| PARP12 | PARP-12/ARTD12 | Poly(ADP-Ribose) Polymerase Family Member 12 | [45][165][167] | |
| PARP14 | PARP-14 | Poly(ADP-Ribose) Polymerase Family Member 14 | [165] | |
| PARP15 | PARP-15 | Poly(ADP-Ribose) Polymerase Family Member 15 | [165] | |
| PATL1 | PATL1 | PAT1 Homolog 1, Processing Body mRNA Decay Factor | [44][45] | yes[45] |
| PAWR | PAWR | PRKC apoptosis WT1 regulator protein | [25] | |
| PCBP1 | PCBP1/HNRNPE1 | Poly(RC) Binding Protein 1 | [44][45] | |
| PCBP2 | PCBP2/HNRNPE2 | Poly(RC) Binding Protein 2 | [25][44][45][73] | |
| PCNA | PCNA | Proliferating cell nuclear antigen | [25] | |
| PDAP1 | PDAP1 | PDGFA Associated Protein 1 | [44] | |
| PDCD4 | PDCD4 | Programmed Cell Death 4 | [168] | |
| PDCD6IP | PDCD6IP | Programmed cell death 6-interacting protein | [25] | |
| PDIA3 | PDIA3 | Protein Disulfide Isomerase Family A Member 3 | [44] | |
| PDLIM1 | PDLIM1 | PDZ and LIM domain protein 1 | [25] | |
| PDLIM4 | PDLIM4 | PDZ and LIM domain protein 4 | [25] | |
| PDLIM5 | PDLIM5 | PDZ and LIM domain protein 5 | [25] | |
| PDS5B | PDS5B | Sister chromatid cohesion protein PDS5 homolog B | [25] | |
| PEF1 | PEF1 | Penta-EF-Hand Domain Containing 1 | [44] | |
| PEG10 | PEG10 | Paternally Expressed 10 | [45] | |
| PELO | PELO | Protein pelota homolog | [25] | |
| PEPD | Peptidase D | Peptidase D | [44] | |
| PEX11B | PEX11B | Peroxisomal Biogenesis Factor 11 Beta | [44] | |
| PFDN4 | PFDN4 | Prefoldin subunit 4 | [25] | |
| PFN1 | Profilin 1 | Profilin 1 | [25][169] | |
| PFN2 | Profilin 2 | Profilin 2 | [25][169] | |
| PGAM5 | PGAM5 | Serine/threonine-protein phosphatase PGAM5, mitochondrial | [25] | |
| PGP | PGP/G3PP | Phosphoglycolate Phosphatase | [44] | |
| PHB2 | Prohibitin 2 | Prohibitin 2 | [22] | |
| PHLDB2 | PHLDB2 | Pleckstrin homology-like domain family B member 2 | [25] | |
| PKP1 | Plakophilin 1 | Plakophilin 1 | [117] | |
| PKP2 | Plakophilin 2 | Plakophilin 2 | [25] | |
| PKP3 | Plakophilin 3 | Plakophilin 3 | [117] | |
| PNPT1 | PNPase I | Polyribonucleotide Nucleotidyltransferase 1 | [44] | |
| POLR2B | POLR2B | DNA-directed RNA polymerase | [25][56] | |
| POM121 | POM121 | POM121 Transmembrane Nucleoporin | [122] | |
| POP7 | RPP20 | POP7 Homolog, Ribonuclease P/MRP Subunit | [170] | |
| PPME1 | PPME1 | Protein phosphatase methylesterase 1 | [25] | |
| PPP1R8 | PPP1R8 | Protein Phosphatase 1 Regulatory Subunit 8 | [44] | |
| PPP1R10 | PPP1R10 | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | [25][56] | |
| PPP1R18 | PPP1R18 | Phostensin | [25] | |
| PPP2R1A | PPP2R1A | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | [25][56] | |
| PPP2R1B | PPP2R1B | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform | [44] | |
| PQBP1 | PQBP-1 | Polyglutamine Binding Protein 1 | ||
| PRDX1 | PRDX1 | Peroxiredoxin-1 | [25][44] | |
| PRDX6 | PRDX6 | Peroxiredoxin-6 | [25] | |
| PRKAA2 | AMPK-a2 | Protein Kinase AMP-Activated Catalytic Subunit Alpha 2 | [21] | |
| PRKCA | PKC-ɑ | Protein Kinase C Alpha | [118] | |
| PRKRA | PACT | Protein Activator Of Interferon Induced Protein Kinase EIF2AK2 | [25][54] | |
| PRMT1 | PRMT1 | Protein arginine N-methyltransferase 1 | [25] | |
| PRMT5 | PRMT5 | Protein arginine N-methyltransferase 5 | [25] | |
| PRRC2A | PRRC2A | Proline Rich Coiled-Coil 2A | [25][44][45] | |
| PRRC2B | PRRC2B | Proline Rich Coiled-Coil 2B | [44][45] | |
| PRRC2C | PRRC2C | Proline Rich Coiled-Coil 2C | [25][44][45][56] | |
| PSMD2 | PSMD2 | 26S proteasome non-ATPase regulatory subunit 2 | [25][171] | |
| PSPC1 | PSP1 | Paraspeckle Component 1 | [44] | |
| PTBP1 | PTBP1 | Polypyrimidine tract-binding protein 1 | [44] | |
| PTBP3 | PTBP3 | Polypyrimidine tract-binding protein 3 | [25][44][45] | |
| PTGES3 | PTGES3 | Prostaglandin E synthase 3 | [25] | |
| PTK2 | FAK | Protein Tyrosine Kinase 2 | [123] | |
| PUM1 | Pumilio-1 | Pumilio homolog 1 | [25][44][45] | yes[35] |
| PUM2 | Pumilio-2 | Pumilio RNA Binding Family Member 2 | [44][45][66] | |
| PURA | PURA | Transcriptional activator protein Pur-alpha | [25][44][172][173] | |
| PURB | PURB | Transcriptional activator protein Pur-beta | [25][44] | |
| PWP1 | PWP1 | PWP1 Homolog, Endonuclein | [44] | |
| PXDNL | PMR1 | Peroxidasin Like | [174] | |
| PYCR1 | PYCR1 | Pyrroline-5-carboxylate reductase | [25] | |
| QKI | QKI/HQK | QKI, KH Domain Containing RNA Binding | [44] | |
| R3HDM1 | R3HDM1 | R3H Domain Containing 1 | [44][45] | |
| R3HDM2 | R3HDM2 | R3H Domain Containing 2 | [45] | |
| RAB1A | RAB1A | Ras-related protein Rab-1A | [25][56] | |
| RACGAP1 | RACGAP1 | Rac GTPase-activating protein 1 | [25] | |
| RACK1 | RACK1 | Receptor For Activated C Kinase 1 | [22][105][175] | |
| RAD21 | RAD21 | Double-strand-break repair protein rad21 homolog | [25] | |
| RAE1 | RAE1 | Ribonucleic Acid Export 1 | [122] | |
| RAN | RAN | RAN, Member RAS Oncogene Family | [144][122] | |
| RANBP1 | RANBP1 | Ran-specific GTPase-activating protein | [25] | |
| RANBP2 | RANBP2/NUP358 | RAN Binding Protein 2 | [122] | |
| RBBP4 | RBBP4 | Histone-binding protein RBBP4 | [25] | |
| RBFOX1 | RBFOX1 | RNA binding protein fox-1 homolog | [25][176][177] | yes[177] |
| RBFOX2 | RBFOX2 | RNA binding protein fox-1 homolog 2 | [176] | |
| RBFOX3 | RBFOX3 | RNA binding protein fox-1 homolog 3 | [176] | |
| RBM12B | RBM12B | RNA-binding protein 12B | [25] | |
| RBM15 | RBM15 | RNA-binding protein 15 | [44] | |
| RBM17 | RBM17 | RNA-binding protein 17 | [44] | |
| RBM25 | RBM25 | RNA-binding protein 25 | [44] | |
| RBM26 | RBM26 | RNA-binding protein 26 | [25] | |
| RBM3 | RBM3 | RNA-binding protein 3 | [44] | |
| RBM38 | RBM38 | RNA-binding protein 38 | [44] | |
| RBM4 | RBM4 | RNA Binding Motif Protein 4 | [44][178] | |
| RBM4B | RBM4B | RNA Binding Motif Protein 4B | [44] | |
| RBM42 | RBM42 | RNA Binding Motif Protein 42 | [133] | |
| RBM45 | RBM45 | RNA Binding Motif Protein 45 | ||
| RBM47 | RBM47 | RNA Binding Motif Protein 47 | [45] | |
| RBMS1 | RBMS1 | RNA-binding motif, single-stranded-interacting protein 1 | [25][44][45] | |
| RBMS2 | RBMS2 | RNA-binding motif, single-stranded-interacting protein 2 | [25][44][45] | |
| RBMX | RBMX | RNA Binding Motif Protein, X-Linked | [45] | |
| RBPMS | RBPMS | RNA-binding protein with multiple splicing | [179] | |
| RC3H1 | Roquin-1 | Ring Finger And CCCH-Type Domains 1 | [44][45][180] | |
| RC3H2 | MNAB | Ring Finger And CCCH-Type Domains 2 | [45][180] | |
| RCC1 | RCC1 | Regulator of chromosome condensation | [25] | |
| RCC2 | RCC2 | Protein RCC2 | [25] | |
| RECQL | RECQL1 | RecQ Like Helicase | [44] | |
| RFC3 | RFC3 | Replication factor C subunit 3 | [25] | |
| RFC4 | RFC4 | Replication factor C subunit 4 | [25] | |
| RGPD3 | RGPD3 | RanBP2-like and GRIP domain-containing protein 3 | [25] | |
| RHOA | RhoA | Ras Homolog Family Member A | [23] | |
| RNASEL | RNAse L | Ribonuclease L | [83][64] | |
| RNF214 | RNF214 | RING finger protein 214 | [25][44] | |
| RNF219 | RNF219 | RING finger protein 219 | [45] | yes[45] |
| RNF25 | RNF25 | Ring Finger Protein 25 | [44] | |
| RNH1 | RNH1 | Ribonuclease inhibitor | [25][53] | |
| ROCK1 | ROCK1 | Rho Associated Coiled-Coil Containing Protein Kinase 1 | [23] | |
| RPS19 | Ribosomal Protein S19 | Ribosomal Protein S19 | [95] | |
| RPS3 | 40S Ribosomal Protein S3 | 40S Ribosomal Protein S3 | [93][95] | yes[35] |
| RPS6 | Ribosomal Protein S6 | Ribosomal Protein S6 | [63][93][4][100][156] | |
| RPS11 | Ribosomal Protein S11 | Ribosomal Protein S11 | [44] | |
| RPS24 | Ribosomal Protein S24 | Ribosomal Protein S24 | [44] | |
| RPS6KA3 | RSK2 | Ribosomal Protein S6 Kinase A3 | [181] | |
| RPS6KB1 | S6K1 | Ribosomal Protein S6 Kinase B1 | [156] | |
| RPS6KB2 | S6K2 | Ribosomal Protein S6 Kinase B2 | [156] | |
| RPTOR | RAPTOR | Regulatory Associated Protein of mTOR Complex 1 | [85][91][156] | |
| RSL1D1 | RSL1D1 | Ribosomal L1 domain-containing protein 1 | [25] | |
| RTCB | RTCB | tRNA-splicing ligase RtcB homolog, formerly C22orf28 | [25][44] | |
| RTRAF | RTRAF (formerly C14orf166) | RNA Transcription, Translation And Transport Factor | [44] | |
| S100A7A | S100A7A | Protein S100-A7A | [25] | |
| S100A9 | S100A9 | Protein S100-A9 | [25] | yes[35] |
| SAFB2 | SAFB2 | Scaffold attachment factor B2 | [25][44] | yes[35] |
| SAMD4A | SMAUG1 | Sterile Alpha Motif Domain Containing 4A | [182] | |
| SAMD4B | SMAUG2 | Sterile Alpha Motif Domain Containing 4B | [44] | |
| SCAPER | SCAPER | S-Phase Cyclin A Associated Protein In The ER | [45] | |
| SEC24C | SEC24C | Protein transport protein Sec24C | [25][44] | |
| SECISBP2 | SECIS Binding Protein 2 | SECIS Binding Protein 2 | [44][45] | |
| SERBP1 | PAI-RBP1/SERBP1 | SERPINE1 mRNA Binding Protein 1 | [49][183][80] | |
| SERPINE1 | PAI-1/Serpin E1 | Serpine Family E Member 1 | [184] | |
| SF1 | SF1 | Splicing Factor 1 | [44] | |
| SFN | SFN | 14-3-3 protein sigma | [25] | |
| SFPQ | PSF | Splicing Factor Proline And Glutamine Rich | [25][158] | |
| SFRS3 | SFRS3 | Serine/arginine-rich splicing factor 3 | [25] | |
| SIPA1L1 | SIPA1L1 | Signal-induced proliferation-associated 1-like protein 1 | [25] | |
| SIRT6 | Sirtuin 6 | Sirtuin 6 | [185] | |
| SLBP | Stem-Loop Binding Protein | Stem-Loop Binding Protein | [44] | |
| SMAP2 | SMAP2 | Small ArfGAP2 | [45] | |
| SMARCA1 | SMARCA1/SNF2L1 | Probable global transcription activator SNF2L1 | [25] | |
| SMC4 | SMC4 | Structural maintenance of chromosomes protein | [25] | |
| SMG1 | SMG-1 | SMG1, Nonsense Mediated mRNA Decay Associated PI3K Related Kinase | [182][186] | |
| SMG6 | SMG6 | SMG6, Nonsense Mediated mRNA Decay Factor | [45] | |
| SMG7 | SMG7 | SMG7, Nonsense Mediated mRNA Decay Factor | [45] | yes[45] |
| SMN1 | Survival of Motor Neuron | Survival Of Motor Neuron 1, Telomeric | [170][187][188] | |
| SMU1 | SMU1 | WD40 repeat-containing protein SMU1 | [25] | |
| SMYD5 | SMYD5 | SMYD Family Member 5 | [44] | |
| SND1 | Tudor-SN | Staphylococcal Nuclease And Tudor Domain Containing 1 | [44][45][47][189] | |
| SNRPF | SNRPF | Small nuclear ribonucleoprotein F | [25] | |
| SNTB2 | SNTB2 | Beta-2-syntrophin | [25] | |
| SOGA3 | SOGA3 | SOGA Family Member 3 | [44] | |
| SORBS1 | SORBS1 | Sorbin and SH3 domain-containing protein 1 | [25] | |
| SORBS3 | Vinexin | Sorbin And SH3 Domain Containing 3 | [190] | |
| SOX3 | SOX3 | SRY-Box 3 | [44] | |
| SPAG5 | Astrin | Sperm Associated Antigen 5 | [85][156] | |
| SPATS2 | SPATS2/SPATA10/SCR59 | Spermatogenesis Associated Serine Rich 2 | [44] | |
| SPATS2L | SGNP | Spermatogenesis Associated Serine Rich 2 Like | [25][191] | |
| SPECC1L | SPECC1L | Cytospin-A | [25] | |
| SQSTM1 | SQSTM1/p62 | Sequestosome 1 | [59] | |
| SRI | SRI | Sorcin | [25][44] | |
| SRP68 | Signal Recognition Particle 68 | Signal Recognition Particle 68 | [44][48] | |
| SRP9 | SRP9 | Signal Recognition Particle 9 | [192] | |
| SRRT | SRRT | Serrate RNA effector molecule homolog | [25] | |
| SRSF1 | ASF/SF2 | Serine And Arginine Rich Splicing Factor 1 | [44][193] | |
| SRSF3 | SRp20 | Serine And Arginine Rich Splicing Factor 3 | [194][195][196][56] | |
| SRSF4 | SRSF4 | Serine/arginine-rich splicing factor 4 | [25] | |
| SRSF5 | SRSF5/SRP40 | Serine/arginine-rich splicing factor 5 | [44] | |
| SRSF7 | 9G8 | Serine And Arginine Rich Splicing Factor 7 | [49] | |
| SRSF9 | SRSF9/SRP30C | Serine/arginine-rich splicing factor 9 | [44] | |
| SS18L1 | SS18L1/CREST | SS18L1, nBAF Chromatin Remodeling Complex Subunit | [197] | |
| ST7 | ST7/FAM4A1/HELG/RAY1/TSG7 | Suppression Of Tumorigenicity 7 | [45] | yes[45] |
| STAT1 | STAT1 | Signal transducer and activator of transcription 1-alpha/beta | [25] | |
| STAU1 | Staufen 1 | Staufen Double-Stranded RNA Binding Protein 1 | [25][44][107][66][198] | |
| STAU2 | Staufen 2 | Staufen Double-Stranded RNA Binding Protein 2 | [25][44][45][107] | yes[35] |
| STIP1 | STIP1/HOP | Stress-induced-phosphoprotein 1 | [25][54] | |
| STRAP | STRAP | Serine-threonine kinase receptor-associated protein | [25][44] | |
| SUGP2 | SUGP2 | SURP and G-patch domain-containing protein 2 | [25] | |
| SUGT1 | SUGT1 | SGT1 Homolog, MIS12 Kinetochore Complex Assembly Cochaperone | [45] | |
| SUN1 | SUN1 | SUN domain-containing protein 1 | [25] | |
| SYCP3 | SYCP3 | Synaptonemal complex protein 3 | [25] | |
| SYK | SYK | Spleen Associated Tyrosine Kinase | [124] | |
| SYNCRIP | SYNCRIP | Heterogeneous nuclear ribonucleoprotein Q | [25][44][45][199] | yes[35] |
| TAGLN3 | Transgelin 3 | Transgelin 3 | [44] | |
| TAF15 | TAF15 | TATA-Box Binding Protein Associated Factor 15 | [25][44][112][113][200][56] | |
| TARDBP | TDP-43 | TAR DNA Binding Protein | [25][108][201][202][129][132][98][203][204][205] | |
| TBRG1 | TBRG1 | Transforming Growth Factor Beta Regulator 1 | [44] | |
| TCEA1 | TCEA1 | Transcription elongation factor A protein 1 | [25] | |
| TCP1 | TCP1 | T-complex protein 1 subunit alpha | [25] | |
| TDRD3 | Tudor Domain Containing 3 | Tudor Domain Containing 3 | [44][45][80][206][207][208] | |
| TDRD7 | Tudor Domain Containing 7 | Tudor Domain Containing 7 | [45] | |
| TERT | TERT | Telomerase Reverse Transcriptase | [209] | |
| THOC2 | THOC2 | THO Complex 2 | [122] | |
| THRAP3 | THRAP3 | Thyroid Hormone Receptor Associated Protein 3 | [44] | |
| TIA1 | TIA-1 | TIA1 Cytotoxic Granule Associated RNA Binding Protein | [4][25][44][49][84][34][66][75][90][116][127][128][163][187][204][210][56] | |
| TIAL1 | TIAR | TIA1 Cytotoxic Granule Associated RNA Binding Protein Like 1 | [25][44][45][66][100][107][108][163][211][187][197] | |
| TMEM131 | TMEM131 | Transmembrane Protein 131 | [45] | yes[45] |
| TMOD3 | TMOD3 | Tropomodulin-3 | [25] | |
| TNKS | PARP-5a | Tankyrase | [165] | |
| TNKS1BP1 | TNKS1BP1 | 182 kDa tankyrase-1-binding protein | [25][45] | yes[45] |
| TNPO1 | Transportin-1 | Transportin-1/Karyopherin (Importin) Beta 2 | [25][44][122][212][213] | |
| TNPO2 | Transportin-2 | Transportin-2 | [25][45] | |
| TNRC6A | TNRC6A | Trinucleotide repeat-containing gene 6A protein | [44][45] | yes[45] |
| TNRC6B | TNRC6B | Trinucleotide repeat-containing gene 6B protein | [25][44][45] | yes[45] |
| TNRC6C | TNRC6C | Trinucleotide repeat-containing gene 6C protein | [44][45] | yes[45] |
| TOMM34 | TOMM34 | Mitochondrial import receptor subunit TOM34 | [25] | |
| TOP3B | Topoisomerase (DNA) III Beta | Topoisomerase (DNA) III Beta | [45][207][214] | |
| TPM1 | TPM1 | Tropomyosin alpha-1 chain | [25] | |
| TPM2 | TPM2 | Tropomyosin beta chain | [25] | |
| TPR | TPR | Translocated Promoter Region, Nuclear Basket Protein | [122] | |
| TRA2B | TRA2B | Transformer 2 Beta Homolog | [45] | |
| TRAF2 | TRAF2 | TNF Receptor Associated Factor 2 | [104] | |
| TRDMT1 | DNMT2 | tRNA Aspartic Acid Methyltransferase 1 | [215] | |
| TRIM21 | TRIM21 | E3 ubiquitin-protein ligase TRIM21 | [25] | |
| TRIM25 | TRIM25 | E3 ubiquitin/ISG15 ligase TRIM25 | [25][44][56] | |
| TRIM56 | TRIM56 | E3 ubiquitin-protein ligase TRIM56 | [25][45][56] | |
| TRIM71 | TRIM71 | E3 ubiquitin-protein ligase TRIM71 | [44] | |
| TRIP6 | TRIP6 | Thyroid receptor-interacting protein 6 | [25][44] | |
| TROVE2 | RORNP | TROVE Domain Family Member 2 | [44] | |
| TTC17 | TTC17 | Tetratricopeptide Repeat Domain 17 | [45] | yes[45] |
| TUBA1C | TUBA1C | Tubulin alpha-1C chain | [25] | |
| TUBA3C | TUBA3C | Tubulin alpha-3C/D chain | [25] | |
| TUBA4A | TUBA4A | Tubulin alpha-4A chain | [25] | |
| TUBB3 | TUBB3 | Tubulin beta-3 chain | [25] | |
| TUBB8 | TUBB8 | Tubulin beta-8 chain | [25] | |
| TUFM | TUFM | Elongation factor Tu, mitochondrial | [25] | |
| TXN | TXN | Thioredoxin | [25] | |
| TXNDC17 | TXNDC17 | Thioredoxin Domain Containing 17 | [44] | |
| U2AF1 | U2AF1 | Splicing factor U2AF 35 kDa subunit | [25] | |
| UBA1 | UBA1 | Ubiquitin-like modifier-activating enzyme 1 | [25] | |
| UBAP2 | UBAP2 | Ubiquitin-associated protein 2 | [25][44][45][56] | |
| UBAP2L | UBAP2L | Ubiquitin-associated protein 2-like | [25][44][45][216][217][56] | |
| UBB | Ubiquitin | Ubiquitin | [109][127] | |
| UBL5 | Ubiquitin Like 5 | Ubiquitin Like 5 | [44] | |
| UBQLN2 | Ubiquilin 2 | Ubiquilin 2 | [218] | |
| ULK1 | ULK1 | Unc-51 Like Autophagy Activating Kinase 1 | [219] | |
| ULK2 | ULK2 | Unc-51 Like Autophagy Activating Kinase 2 | [219] | |
| UPF1 | UPF1 | UPF1, RNA Helicase and ATPase | [25][44][45][186][56] | yes[35] |
| UPF2 | UPF2 | UPF2, RNA Helicase and ATPase | [186] | |
| UPF3B | UPF3B | UPF3B, Regulator of Nonsense Mediated mRNA Decay | [44] | |
| USP10 | USP10 | Ubiquitin Specific Peptidase 10 | [25][44][45][63][34][175][56] | |
| USP11 | USP11 | Ubiquitin Specific Peptidase 11 | [44] | |
| USP13 | USP13 | Ubiquitin Specific Peptidase 13 | [220] | |
| USP5 | USP5 | Ubiquitin carboxyl-terminal hydrolase 5 | [25][220] | |
| USP9X | USP9X | Ubiquitin Specific Peptidase 9, X-Linked | [208] | |
| UTP18 | UTP18 | UTP18, Small Subunit Processome Component | [44] | |
| VASP | VASP | Vasodilator-stimulated phosphoprotein | [25] | |
| VBP1 | VBP1 | VHL Binding Protein 1 | [44] | |
| VCP | VCP | Valosin Containing Protein | [25][221][171][219] | |
| WBP2 | WBP2 | WW Domain Binding Protein 2 | [44] | |
| WDR47 | WDR47 | WD Repeat Domain 47 | [44] | |
| WDR62 | WDR62 | WD Repeat Domain 62 | [151] | |
| XPO1 | XPO1/CRM1 | Exportin 1 | [122] | |
| XRN1 | XRN1 | 5'-3' Exoribonuclease 1 | [36][44][45] | yes[36][45] |
| XRN2 | XRN2 | 5'-3' Exoribonuclease 2 | [44] | |
| YARS | YARS | Tyrosine—tRNA ligase, cytoplasmic | [25] | |
| YBX1 | YB-1 | Y-Box Binding Protein 1 | [25][44][49][48][78][89][222] | |
| YBX3 | YBX3/ZONAB | Y-box-binding protein 3 | [25][44][45] | |
| YES1 | YES1 | Tyrosine-protein kinase Yes | [25] | |
| YLPM1 | YLPM1 | YLP Motif Containing 1 | [44] | |
| YTHDF1 | YTHDF1 | YTH domain family protein 1 | [25][44][45][223][224] | |
| YTHDF2 | YTHDF2 | YTH domain family protein 2 | [25][44][45][223][224] | yes[223][224] |
| YTHDF3 | YTHDF3 | YTH domain family protein 3 | [25][32][44][45][223][224] | |
| YWHAB | 14-3-3 | Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein Beta | [25][152] | |
| YWHAH | 14-3-3 | 14-3-3 protein eta | [25] | |
| YWHAQ | 14-3-3 | 14-3-3 protein theta | [25] | |
| ZBP1 | ZBP1 | Z-DNA Binding Protein 1 | [225][226] | |
| ZCCHC11 | ZCCHC11 | Zinc finger CCCH domain-containing protein 11 | [45] | |
| ZCCHC14 | ZCCHC14 | Zinc finger CCCH domain-containing protein 14 | [45] | |
| ZC3H11A | ZC3H11A | Zinc finger CCCH domain-containing protein 11a | [44] | |
| ZC3H14 | ZC3H14 | Zinc finger CCCH domain-containing protein 14 | [25] | |
| ZCCHC2 | ZCCHC2 | Zinc finger CCCH domain-containing protein 2 | [45] | |
| ZCCHC3 | ZCCHC3 | Zinc finger CCCH domain-containing protein 3 | [45] | |
| ZC3H7A | ZC3H7A | Zinc finger CCCH domain-containing protein 7A | [25] | |
| ZC3H7B | ZC3H7B | Zinc finger CCCH domain-containing protein 7B | [25][44] | |
| ZC3HAV1 | PARP-13.1/PARP-13.2/ARTD13 | Zinc Finger CCCH-Type Containing, Antiviral 1 | [25][45][165] | yes[35] |
| ZFAND1 | ZFAND1 | Zinc Finger AN1-Type Containing 1 | [171] | |
| ZFP36 | TTP/TIS11 | ZFP36 Ring Finger Protein/Trisetrapolin | [36][44][151][227][228][229] | yes[36] |
| ZNF598 | ZNF598 | Zinc finger protein 598 | [45] | |
| ZNF638 | ZNF638 | Zinc finger protein 638 | [25] |
References
- ↑ "Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis". The Plant Cell 27 (3): 926–943. March 2015. doi:10.1105/tpc.114.134494. PMID 25736060.
- ↑ Hirose, Tetsuro; Ninomiya, Kensuke; Nakagawa, Shinichi; Yamazaki, Tomohiro (2022-11-23). "A guide to membraneless organelles and their various roles in gene regulation" (in en). Nature Reviews Molecular Cell Biology 24 (4): 288–304. doi:10.1038/s41580-022-00558-8. ISSN 1471-0080. PMID 36424481. https://www.nature.com/articles/s41580-022-00558-8.
- ↑ Hirose, Tetsuro; Ninomiya, Kensuke; Nakagawa, Shinichi; Yamazaki, Tomohiro (April 2023). "A guide to membraneless organelles and their various roles in gene regulation" (in en). Nature Reviews Molecular Cell Biology 24 (4): 288–304. doi:10.1038/s41580-022-00558-8. ISSN 1471-0080. PMID 36424481. https://www.nature.com/articles/s41580-022-00558-8.
- ↑ 4.0 4.1 4.2 4.3 4.4 "Prolonged translation arrest in reperfused hippocampal cornu Ammonis 1 is mediated by stress granules". Neuroscience 134 (4): 1223–1245. 2005. doi:10.1016/j.neuroscience.2005.05.047. PMID 16055272.
- ↑ "Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs". Molecular and Cellular Biology 9 (3): 1298–1308. March 1989. doi:10.1128/mcb.9.3.1298. PMID 2725500.
- ↑ Paul J. Anderson, Brigham and Women's Hospital
- ↑ "Translationally repressed mRNA transiently cycles through stress granules during stress". Molecular Biology of the Cell 19 (10): 4469–4479. October 2008. doi:10.1091/mbc.E08-05-0499. PMID 18632980.
- ↑ 8.0 8.1 8.2 8.3 "The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules". Molecular Cell 68 (4): 808–820.e5. November 2017. doi:10.1016/j.molcel.2017.10.015. PMID 29129640.
- ↑ "mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction". The Journal of Cell Biology 217 (12): 4124–4140. December 2018. doi:10.1083/jcb.201806183. PMID 30322972.
- ↑ "Isolation of mammalian stress granule cores for RNA-Seq analysis". Methods 137: 49–54. March 2018. doi:10.1016/j.ymeth.2017.11.012. PMID 29196162.
- ↑ "Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo". The Plant Cell 9 (12): 2171–2181. December 1997. doi:10.1105/tpc.9.12.2171. PMID 9437862.
- ↑ "Cytosolic heat-stress proteins Hsp17.7 class I and Hsp17.3 class II of tomato act as molecular chaperones in vivo". Planta 211 (4): 575–582. September 2000. doi:10.1007/s004250000315. PMID 11030557.
- ↑ "Messenger RNA-binding properties of nonpolysomal ribonucleoproteins from heat-stressed tomato cells". Plant Physiology 120 (1): 23–32. May 1999. doi:10.1104/pp.120.1.23. PMID 10318680.
- ↑ "The prosome: an ubiquitous morphologically distinct RNP particle associated with repressed mRNPs and containing specific ScRNA and a characteristic set of proteins". The EMBO Journal 3 (1): 29–34. January 1984. doi:10.1002/j.1460-2075.1984.tb01757.x. PMID 6200323.
- ↑ "Nitric oxide triggers the assembly of "type II" stress granules linked to decreased cell viability". Cell Death & Disease 9 (11): 1129. November 2018. doi:10.1038/s41419-018-1173-x. PMID 30425239.
- ↑ "A Systems-Level Study Reveals Regulators of Membrane-less Organelles in Human Cells". Molecular Cell 72 (6): 1035–1049.e5. December 2018. doi:10.1016/j.molcel.2018.10.036. PMID 30503769.
- ↑ 17.0 17.1 17.2 17.3 "Stress-specific differences in assembly and composition of stress granules and related foci". Journal of Cell Science 130 (5): 927–937. March 2017. doi:10.1242/jcs.199240. PMID 28096475.
- ↑ Qifti, Androniqi; Jackson, Lela; Singla, Ashima; Garwain, Osama; Scarlata, Suzanne (2021-10-19). "Stimulation of phospholipase Cβ1 by Gα q promotes the assembly of stress granule proteins" (in en). Science Signaling 14 (705). doi:10.1126/scisignal.aav1012. ISSN 1945-0877. https://www.science.org/doi/10.1126/scisignal.aav1012.
- ↑ "Stress granule assembly is mediated by prion-like aggregation of TIA-1". Molecular Biology of the Cell 15 (12): 5383–5398. December 2004. doi:10.1091/mbc.E04-08-0715. PMID 15371533.
- ↑ "Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation". Experimental Cell Research 290 (2): 227–233. November 2003. doi:10.1016/S0014-4827(03)00290-8. PMID 14567982.
- ↑ 21.0 21.1 "5'-AMP-activated protein kinase alpha regulates stress granule biogenesis". Biochimica et Biophysica Acta 1853 (7): 1725–1737. July 2015. doi:10.1016/j.bbamcr.2015.03.015. PMID 25840010.
- ↑ 22.0 22.1 22.2 "A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly". Nature Cell Biology 10 (10): 1224–1231. October 2008. doi:10.1038/ncb1783. PMID 18794846.
- ↑ 23.0 23.1 23.2 23.3 "RhoA/ROCK1 signaling regulates stress granule formation and apoptosis". Cellular Signalling 22 (4): 668–675. April 2010. doi:10.1016/j.cellsig.2009.12.001. PMID 20004716.
- ↑ 24.0 24.1 "RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome". Proceedings of the National Academy of Sciences of the United States of America 115 (11): 2734–2739. March 2018. doi:10.1073/pnas.1800038115. PMID 29483269. Bibcode: 2018PNAS..115.2734V.
- ↑ 25.000 25.001 25.002 25.003 25.004 25.005 25.006 25.007 25.008 25.009 25.010 25.011 25.012 25.013 25.014 25.015 25.016 25.017 25.018 25.019 25.020 25.021 25.022 25.023 25.024 25.025 25.026 25.027 25.028 25.029 25.030 25.031 25.032 25.033 25.034 25.035 25.036 25.037 25.038 25.039 25.040 25.041 25.042 25.043 25.044 25.045 25.046 25.047 25.048 25.049 25.050 25.051 25.052 25.053 25.054 25.055 25.056 25.057 25.058 25.059 25.060 25.061 25.062 25.063 25.064 25.065 25.066 25.067 25.068 25.069 25.070 25.071 25.072 25.073 25.074 25.075 25.076 25.077 25.078 25.079 25.080 25.081 25.082 25.083 25.084 25.085 25.086 25.087 25.088 25.089 25.090 25.091 25.092 25.093 25.094 25.095 25.096 25.097 25.098 25.099 25.100 25.101 25.102 25.103 25.104 25.105 25.106 25.107 25.108 25.109 25.110 25.111 25.112 25.113 25.114 25.115 25.116 25.117 25.118 25.119 25.120 25.121 25.122 25.123 25.124 25.125 25.126 25.127 25.128 25.129 25.130 25.131 25.132 25.133 25.134 25.135 25.136 25.137 25.138 25.139 25.140 25.141 25.142 25.143 25.144 25.145 25.146 25.147 25.148 25.149 25.150 25.151 25.152 25.153 25.154 25.155 25.156 25.157 25.158 25.159 25.160 25.161 25.162 25.163 25.164 25.165 25.166 25.167 25.168 25.169 25.170 25.171 25.172 25.173 25.174 25.175 25.176 25.177 25.178 25.179 25.180 25.181 25.182 25.183 25.184 25.185 25.186 25.187 25.188 25.189 25.190 25.191 25.192 25.193 25.194 25.195 25.196 25.197 25.198 25.199 25.200 25.201 25.202 25.203 25.204 25.205 25.206 25.207 25.208 25.209 25.210 25.211 25.212 25.213 25.214 25.215 25.216 25.217 25.218 25.219 25.220 25.221 25.222 25.223 25.224 25.225 25.226 25.227 25.228 25.229 25.230 25.231 25.232 25.233 25.234 25.235 25.236 25.237 25.238 25.239 25.240 25.241 25.242 25.243 25.244 25.245 25.246 25.247 25.248 25.249 25.250 25.251 25.252 25.253 25.254 25.255 25.256 25.257 25.258 25.259 25.260 25.261 25.262 25.263 25.264 25.265 25.266 25.267 25.268 25.269 25.270 25.271 25.272 25.273 25.274 25.275 25.276 25.277 25.278 25.279 25.280 25.281 25.282 25.283 25.284 25.285 25.286 25.287 25.288 25.289 25.290 25.291 25.292 25.293 25.294 25.295 25.296 25.297 25.298 25.299 25.300 25.301 25.302 25.303 25.304 25.305 25.306 25.307 25.308 25.309 25.310 25.311 25.312 25.313 25.314 25.315 25.316 25.317 25.318 25.319 25.320 25.321 25.322 25.323 25.324 25.325 25.326 25.327 25.328 "ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure". Cell 164 (3): 487–498. January 2016. doi:10.1016/j.cell.2015.12.038. PMID 26777405.
- ↑ 26.0 26.1 26.2 "Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain". The Journal of Biological Chemistry 283 (50): 35186–35198. December 2008. doi:10.1074/jbc.M804857200. PMID 18854321.
- ↑ "The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex". Molecular Cell 43 (6): 962–972. September 2011. doi:10.1016/j.molcel.2011.08.008. PMID 21925384.
- ↑ "Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis". Journal of Molecular Biology 427 (9): 1779–1796. May 2015. doi:10.1016/j.jmb.2015.02.015. PMID 25724843.
- ↑ 29.0 29.1 "Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation". Scientific Reports 6 (1): 25996. May 2016. doi:10.1038/srep25996. PMID 27180681. Bibcode: 2016NatSR...625996V.
- ↑ "Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies". Cell 174 (4): 791–802. August 2018. doi:10.1016/j.cell.2018.07.023. PMID 30096311.
- ↑ "Spatial Organization of Single mRNPs at Different Stages of the Gene Expression Pathway". Molecular Cell 72 (4): 727–738.e5. November 2018. doi:10.1016/j.molcel.2018.10.010. PMID 30415950.
- ↑ 32.0 32.1 "Dynamic m6A methylation facilitates mRNA triaging to stress granules". Life Science Alliance 1 (4): e201800113. August 2018. doi:10.26508/lsa.201800113. PMID 30456371.
- ↑ "Modulation of RNA Condensation by the DEAD-Box Protein eIF4A". Cell 180 (3): 411–426.e16. February 2020. doi:10.1016/j.cell.2019.12.031. PMID 31928844.
- ↑ 34.0 34.1 34.2 34.3 34.4 34.5 34.6 "G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA". The Journal of Cell Biology 209 (1): 73–84. April 2015. doi:10.1083/jcb.201408092. PMID 25847539.
- ↑ 35.00 35.01 35.02 35.03 35.04 35.05 35.06 35.07 35.08 35.09 35.10 35.11 35.12 35.13 35.14 35.15 35.16 35.17 35.18 35.19 35.20 35.21 35.22 35.23 35.24 35.25 35.26 35.27 35.28 35.29 35.30 35.31 35.32 35.33 35.34 35.35 35.36 "P-Body Purification Reveals the Condensation of Repressed mRNA Regulons". Molecular Cell 68 (1): 144–157.e5. October 2017. doi:10.1016/j.molcel.2017.09.003. PMID 28965817.
- ↑ 36.00 36.01 36.02 36.03 36.04 36.05 36.06 36.07 36.08 36.09 36.10 36.11 36.12 "Stress granules and processing bodies are dynamically linked sites of mRNP remodeling". The Journal of Cell Biology 169 (6): 871–884. June 2005. doi:10.1083/jcb.200502088. PMID 15967811.
- ↑ "P bodies promote stress granule assembly in Saccharomyces cerevisiae". The Journal of Cell Biology 183 (3): 441–455. November 2008. doi:10.1083/jcb.200807043. PMID 18981231.
- ↑ 38.0 38.1 38.2 Figley MD (2015). Profilin 1, stress granules, and ALS pathogenesis (PhD). Stanford University.
- ↑ 39.0 39.1 "Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS?". Frontiers in Cellular Neuroscience 9: 423. 2015. doi:10.3389/fncel.2015.00423. PMID 26557057.
- ↑ 40.0 40.1 "Properties of Stress Granule and P-Body Proteomes" (in English). Molecular Cell 76 (2): 286–294. October 2019. doi:10.1016/j.molcel.2019.09.014. PMID 31626750.
- ↑ "Methods to Classify Cytoplasmic Foci as Mammalian Stress Granules". Journal of Visualized Experiments (123). May 2017. doi:10.3791/55656. PMID 28570526.
- ↑ "Distinct stages in stress granule assembly and disassembly". eLife 5. September 2016. doi:10.7554/eLife.18413. PMID 27602576.
- ↑ "Isolation of yeast and mammalian stress granule cores". Methods 126: 12–17. August 2017. doi:10.1016/j.ymeth.2017.04.020. PMID 28457979.
- ↑ 44.000 44.001 44.002 44.003 44.004 44.005 44.006 44.007 44.008 44.009 44.010 44.011 44.012 44.013 44.014 44.015 44.016 44.017 44.018 44.019 44.020 44.021 44.022 44.023 44.024 44.025 44.026 44.027 44.028 44.029 44.030 44.031 44.032 44.033 44.034 44.035 44.036 44.037 44.038 44.039 44.040 44.041 44.042 44.043 44.044 44.045 44.046 44.047 44.048 44.049 44.050 44.051 44.052 44.053 44.054 44.055 44.056 44.057 44.058 44.059 44.060 44.061 44.062 44.063 44.064 44.065 44.066 44.067 44.068 44.069 44.070 44.071 44.072 44.073 44.074 44.075 44.076 44.077 44.078 44.079 44.080 44.081 44.082 44.083 44.084 44.085 44.086 44.087 44.088 44.089 44.090 44.091 44.092 44.093 44.094 44.095 44.096 44.097 44.098 44.099 44.100 44.101 44.102 44.103 44.104 44.105 44.106 44.107 44.108 44.109 44.110 44.111 44.112 44.113 44.114 44.115 44.116 44.117 44.118 44.119 44.120 44.121 44.122 44.123 44.124 44.125 44.126 44.127 44.128 44.129 44.130 44.131 44.132 44.133 44.134 44.135 44.136 44.137 44.138 44.139 44.140 44.141 44.142 44.143 44.144 44.145 44.146 44.147 44.148 44.149 44.150 44.151 44.152 44.153 44.154 44.155 44.156 44.157 44.158 44.159 44.160 44.161 44.162 44.163 44.164 44.165 44.166 44.167 44.168 44.169 44.170 44.171 44.172 44.173 44.174 44.175 44.176 44.177 44.178 44.179 44.180 44.181 44.182 44.183 44.184 44.185 44.186 44.187 44.188 44.189 44.190 44.191 44.192 44.193 44.194 44.195 44.196 44.197 44.198 44.199 44.200 44.201 44.202 44.203 44.204 44.205 44.206 44.207 44.208 44.209 44.210 44.211 44.212 44.213 44.214 44.215 44.216 44.217 44.218 44.219 44.220 44.221 44.222 44.223 44.224 44.225 44.226 44.227 44.228 44.229 44.230 44.231 44.232 44.233 44.234 44.235 44.236 44.237 44.238 44.239 44.240 44.241 44.242 44.243 44.244 44.245 44.246 44.247 44.248 44.249 44.250 44.251 44.252 44.253 44.254 44.255 44.256 44.257 "Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules" (in English). Cell 172 (3): 590–604.e13. January 2018. doi:10.1016/j.cell.2017.12.032. PMID 29373831.
- ↑ 45.000 45.001 45.002 45.003 45.004 45.005 45.006 45.007 45.008 45.009 45.010 45.011 45.012 45.013 45.014 45.015 45.016 45.017 45.018 45.019 45.020 45.021 45.022 45.023 45.024 45.025 45.026 45.027 45.028 45.029 45.030 45.031 45.032 45.033 45.034 45.035 45.036 45.037 45.038 45.039 45.040 45.041 45.042 45.043 45.044 45.045 45.046 45.047 45.048 45.049 45.050 45.051 45.052 45.053 45.054 45.055 45.056 45.057 45.058 45.059 45.060 45.061 45.062 45.063 45.064 45.065 45.066 45.067 45.068 45.069 45.070 45.071 45.072 45.073 45.074 45.075 45.076 45.077 45.078 45.079 45.080 45.081 45.082 45.083 45.084 45.085 45.086 45.087 45.088 45.089 45.090 45.091 45.092 45.093 45.094 45.095 45.096 45.097 45.098 45.099 45.100 45.101 45.102 45.103 45.104 45.105 45.106 45.107 45.108 45.109 45.110 45.111 45.112 45.113 45.114 45.115 45.116 45.117 45.118 45.119 45.120 45.121 45.122 45.123 45.124 45.125 45.126 45.127 45.128 45.129 45.130 45.131 45.132 45.133 45.134 45.135 45.136 45.137 45.138 45.139 45.140 45.141 45.142 45.143 45.144 45.145 45.146 45.147 45.148 45.149 45.150 45.151 45.152 45.153 45.154 45.155 45.156 45.157 45.158 45.159 45.160 45.161 45.162 45.163 45.164 45.165 45.166 45.167 45.168 45.169 45.170 45.171 45.172 45.173 45.174 "High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies" (in English). Molecular Cell 69 (3): 517–532.e11. February 2018. doi:10.1016/j.molcel.2017.12.020. PMID 29395067.
- ↑ 46.00 46.01 46.02 46.03 46.04 46.05 46.06 46.07 46.08 46.09 46.10 46.11 46.12 46.13 46.14 46.15 46.16 46.17 46.18 46.19 46.20 46.21 46.22 46.23 46.24 46.25 46.26 46.27 46.28 46.29 46.30 46.31 46.32 46.33 46.34 46.35 46.36 46.37 46.38 46.39 46.40 46.41 46.42 46.43 46.44 46.45 46.46 46.47 46.48 46.49 46.50 46.51 46.52 46.53 46.54 46.55 46.56 46.57 46.58 46.59 46.60 46.61 46.62 46.63 46.64 46.65 46.66 46.67 46.68 46.69 46.70 46.71 46.72 46.73 46.74 46.75 46.76 "Spatiotemporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis" (in English). Molecular Cell 80 (5): 876–891.e6. December 2020. doi:10.1016/j.molcel.2020.10.032. PMID 33217318.
- ↑ 47.0 47.1 "Tudor-SN and ADAR1 are components of cytoplasmic stress granules". RNA 18 (3): 462–471. March 2012. doi:10.1261/rna.027656.111. PMID 22240577.
- ↑ 48.0 48.1 48.2 48.3 48.4 48.5 48.6 "Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules". Journal of Virology 81 (5): 2165–2178. March 2007. doi:10.1128/JVI.02287-06. PMID 17166910.
- ↑ 49.0 49.1 49.2 49.3 49.4 49.5 49.6 49.7 "LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex". Molecular and Cellular Biology 27 (18): 6469–6483. September 2007. doi:10.1128/MCB.00332-07. PMID 17562864.
- ↑ "Cell stress is related to re-localization of Argonaute 2 and to decreased RNA interference in human cells". Nucleic Acids Research 39 (7): 2727–2741. April 2011. doi:10.1093/nar/gkq1216. PMID 21148147.
- ↑ "RNA interference may suppress stress granule formation by preventing argonaute 2 recruitment". American Journal of Physiology. Cell Physiology 316 (1): C81–C91. January 2019. doi:10.1152/ajpcell.00251.2018. PMID 30404558.
- ↑ 52.0 52.1 52.2 52.3 "Microtubule-dependent association of AKAP350A and CCAR1 with RNA stress granules". Experimental Cell Research 315 (3): 542–555. February 2009. doi:10.1016/j.yexcr.2008.11.011. PMID 19073175.
- ↑ 53.0 53.1 "Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival". Journal of Cell Science 126 (Pt 18): 4308–4319. September 2013. doi:10.1242/jcs.134551. PMID 23843625.
- ↑ 54.0 54.1 54.2 54.3 "Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies". Molecular Biology of the Cell 20 (14): 3273–3284. July 2009. doi:10.1091/mbc.E09-01-0082. PMID 19458189.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedKaehler_2012 - ↑ 56.00 56.01 56.02 56.03 56.04 56.05 56.06 56.07 56.08 56.09 56.10 56.11 56.12 56.13 56.14 56.15 56.16 56.17 56.18 56.19 56.20 56.21 56.22 56.23 56.24 56.25 56.26 56.27 56.28 56.29 56.30 56.31 56.32 Cite error: Invalid
<ref>tag; no text was provided for refs named:10 - ↑ "BOULE, a Deleted in Azoospermia Homolog, Is Recruited to Stress Granules in the Mouse Male Germ Cells". PLOS ONE 11 (9): e0163015. 2016. doi:10.1371/journal.pone.0163015. PMID 27632217. Bibcode: 2016PLoSO..1163015K.
- ↑ "C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly, Hypersensitizing Cells to Stress". Molecular Neurobiology 54 (4): 3062–3077. May 2017. doi:10.1007/s12035-016-9850-1. PMID 27037575. https://boris.unibe.ch/80746/.
- ↑ 59.0 59.1 "A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy". Nature Communications 9 (1): 2794. July 2018. doi:10.1038/s41467-018-05273-7. PMID 30022074. Bibcode: 2018NatCo...9.2794C.
- ↑ "Post-translational arginylation of calreticulin: a new isospecies of calreticulin component of stress granules". The Journal of Biological Chemistry 282 (11): 8237–8245. March 2007. doi:10.1074/jbc.M608559200. PMID 17197444.
- ↑ "Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs". Molecular and Cellular Biology 27 (6): 2324–2342. March 2007. doi:10.1128/MCB.02300-06. PMID 17210633.
- ↑ 62.0 62.1 "Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis". Cell Cycle 11 (10): 2006–2021. May 2012. doi:10.4161/cc.20423. PMID 22580459.
- ↑ 63.0 63.1 63.2 "G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits". The Journal of Cell Biology 212 (7): 845–860. March 2016. doi:10.1083/jcb.201508028. PMID 27022092.
- ↑ 64.0 64.1 64.2 "Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1". mBio 6 (2): e02486. March 2015. doi:10.1128/mBio.02486-14. PMID 25784705.
- ↑ 65.0 65.1 "The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly". Journal of Cell Science 120 (Pt 16): 2774–2784. August 2007. doi:10.1242/jcs.009225. PMID 17652158.
- ↑ 66.0 66.1 66.2 66.3 66.4 66.5 66.6 66.7 "Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules". The Journal of Neuroscience 26 (24): 6496–6508. June 2006. doi:10.1523/JNEUROSCI.0649-06.2006. PMID 16775137.
- ↑ "Phosphorylation of hnRNP K by cyclin-dependent kinase 2 controls cytosolic accumulation of TDP-43". Human Molecular Genetics 24 (6): 1655–1669. March 2015. doi:10.1093/hmg/ddu578. PMID 25410660.
- ↑ "Dual localization of the RNA binding protein CUGBP-1 to stress granule and perinucleolar compartment". Experimental Cell Research 314 (3): 543–553. February 2008. doi:10.1016/j.yexcr.2007.10.024. PMID 18164289.
- ↑ "CERKL, a retinal disease gene, encodes an mRNA-binding protein that localizes in compact and untranslated mRNPs associated with microtubules". PLOS ONE 9 (2): e87898. 2014. doi:10.1371/journal.pone.0087898. PMID 24498393. Bibcode: 2014PLoSO...987898F.
- ↑ "The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor". Experimental Cell Research 313 (20): 4130–4144. December 2007. doi:10.1016/j.yexcr.2007.09.017. PMID 17967451.
- ↑ "Yeast Gis2 and its human ortholog CNBP are novel components of stress-induced RNP granules". PLOS ONE 7 (12): e52824. 2012. doi:10.1371/journal.pone.0052824. PMID 23285195. Bibcode: 2012PLoSO...752824R.
- ↑ "Cytoplasmic foci are sites of mRNA decay in human cells". The Journal of Cell Biology 165 (1): 31–40. April 2004. doi:10.1083/jcb.200309008. PMID 15067023.
- ↑ 73.0 73.1 "Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies". RNA 14 (3): 425–431. March 2008. doi:10.1261/rna.780708. PMID 18174314.
- ↑ "Casein Kinase 2 Is Linked to Stress Granule Dynamics through Phosphorylation of the Stress Granule Nucleating Protein G3BP1". Molecular and Cellular Biology 37 (4): e00596–16. February 2017. doi:10.1128/MCB.00596-16. PMID 27920254.
- ↑ 75.0 75.1 75.2 75.3 75.4 "Proline-rich transcript in brain protein induces stress granule formation". Molecular and Cellular Biology 28 (2): 803–813. January 2008. doi:10.1128/MCB.01226-07. PMID 17984221.
- ↑ "DAZL is essential for stress granule formation implicated in germ cell survival upon heat stress". Development 139 (3): 568–578. February 2012. doi:10.1242/dev.075846. PMID 22223682.
- ↑ 77.0 77.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedWilczynska_2005 - ↑ 78.0 78.1 78.2 "MBNL1 associates with YB-1 in cytoplasmic stress granules". Journal of Neuroscience Research 86 (9): 1994–2002. July 2008. doi:10.1002/jnr.21655. PMID 18335541.
- ↑ "DDX3 RNA helicase is required for HIV-1 Tat function". Biochemical and Biophysical Research Communications 441 (3): 607–611. November 2013. doi:10.1016/j.bbrc.2013.10.107. PMID 24183723.
- ↑ 80.0 80.1 80.2 "TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules". Human Molecular Genetics 17 (19): 3055–3074. October 2008. doi:10.1093/hmg/ddn203. PMID 18632687.
- ↑ "Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation". Scientific Reports 6: 25996. May 2016. doi:10.1038/srep25996. PMID 27180681. Bibcode: 2016NatSR...625996V.
- ↑ 82.0 82.1 "Acetylation of intrinsically disordered regions regulates phase separation". Nature Chemical Biology 15 (1): 51–61. January 2019. doi:10.1038/s41589-018-0180-7. PMID 30531905.
- ↑ 83.0 83.1 83.2 83.3 83.4 83.5 "Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity". PLOS ONE 7 (8): e43031. 2012. doi:10.1371/journal.pone.0043031. PMID 22912779. Bibcode: 2012PLoSO...743031O.
- ↑ 84.0 84.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedNonhoff_2007 - ↑ 85.0 85.1 85.2 "Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells". Cell 154 (4): 859–874. August 2013. doi:10.1016/j.cell.2013.07.031. PMID 23953116.
- ↑ 86.0 86.1 86.2 86.3 "Comprehensive Protein Interactome Analysis of a Key RNA Helicase: Detection of Novel Stress Granule Proteins". Biomolecules 5 (3): 1441–1466. July 2015. doi:10.3390/biom5031441. PMID 26184334.
- ↑ "DERA is the human deoxyribose phosphate aldolase and is involved in stress response". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1843 (12): 2913–2925. December 2014. doi:10.1016/j.bbamcr.2014.09.007. PMID 25229427.
- ↑ 88.0 88.1 88.2 88.3 "Dynein and kinesin regulate stress-granule and P-body dynamics". Journal of Cell Science 122 (Pt 21): 3973–3982. November 2009. doi:10.1242/jcs.051383. PMID 19825938.
- ↑ 89.0 89.1 89.2 "The Atypical Dual Specificity Phosphatase hYVH1 Associates with Multiple Ribonucleoprotein Particles". The Journal of Biological Chemistry 292 (2): 539–550. January 2017. doi:10.1074/jbc.M116.715607. PMID 27856639.
- ↑ 90.0 90.1 90.2 "Dynein motor contributes to stress granule dynamics in primary neurons". Neuroscience 159 (2): 647–656. March 2009. doi:10.1016/j.neuroscience.2008.12.053. PMID 19171178.
- ↑ 91.0 91.1 91.2 "Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling". Cell 152 (4): 791–805. February 2013. doi:10.1016/j.cell.2013.01.033. PMID 23415227.
- ↑ "Ribonomic analysis of human DZIP1 reveals its involvement in ribonucleoprotein complexes and stress granules". BMC Molecular Biology 15: 12. July 2014. doi:10.1186/1471-2199-15-12. PMID 24993635.
- ↑ 93.0 93.1 93.2 93.3 93.4 93.5 "Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes". American Journal of Physiology. Cell Physiology 284 (2): C273–C284. February 2003. doi:10.1152/ajpcell.00314.2002. PMID 12388085.
- ↑ 94.0 94.1 "The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses". Journal of Virology 89 (5): 2575–2589. March 2015. doi:10.1128/JVI.02791-14. PMID 25520508.
- ↑ 95.0 95.1 95.2 95.3 95.4 95.5 "Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules". Molecular Biology of the Cell 13 (1): 195–210. January 2002. doi:10.1091/mbc.01-05-0221. PMID 11809833.
- ↑ 96.0 96.1 "eIF5A promotes translation elongation, polysome disassembly and stress granule assembly". PLOS ONE 5 (4): e9942. April 2010. doi:10.1371/journal.pone.0009942. PMID 20376341. Bibcode: 2010PLoSO...5.9942L.
- ↑ 97.0 97.1 "Identification of Neuregulin-2 as a novel stress granule component". BMB Reports 49 (8): 449–454. August 2016. doi:10.5483/BMBRep.2016.49.8.090. PMID 27345716.
- ↑ 98.0 98.1 "Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination". PLOS ONE 7 (6): e38658. 2012. doi:10.1371/journal.pone.0038658. PMID 22761693. Bibcode: 2012PLoSO...738658D.
- ↑ "Sumoylation of eIF4A2 affects stress granule formation". Journal of Cell Science 129 (12): 2407–2415. June 2016. doi:10.1242/jcs.184614. PMID 27160682.
- ↑ 100.0 100.1 100.2 100.3 100.4 100.5 100.6 100.7 100.8 100.9 "Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion". The Journal of Neuroscience 26 (9): 2413–2418. March 2006. doi:10.1523/JNEUROSCI.3680-05.2006. PMID 16510718.
- ↑ 101.0 101.1 101.2 "Inhibition of the ubiquitin-proteasome system induces stress granule formation". Molecular Biology of the Cell 18 (7): 2603–2618. July 2007. doi:10.1091/mbc.E06-12-1079. PMID 17475769.
- ↑ 102.0 102.1 102.2 "Distinct recruitment of human eIF4E isoforms to processing bodies and stress granules". BMC Molecular Biology 17 (1): 21. August 2016. doi:10.1186/s12867-016-0072-x. PMID 27578149.
- ↑ 103.0 103.1 "SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate". The Journal of Biological Chemistry 282 (38): 27953–27959. September 2007. doi:10.1074/jbc.M702317200. PMID 17640873.
- ↑ 104.0 104.1 "Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions". Molecular and Cellular Biology 25 (6): 2450–2462. March 2005. doi:10.1128/MCB.25.6.2450-2462.2005. PMID 15743837.
- ↑ 105.0 105.1 "Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways". Nature Cell Biology 10 (11): 1324–1332. November 2008. doi:10.1038/ncb1791. PMID 18836437.
- ↑ "HuR binding to cytoplasmic mRNA is perturbed by heat shock". Proceedings of the National Academy of Sciences of the United States of America 97 (7): 3073–3078. March 2000. doi:10.1073/pnas.97.7.3073. PMID 10737787. Bibcode: 2000PNAS...97.3073G.
- ↑ 107.0 107.1 107.2 107.3 107.4 "Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes". Molecular Biology of the Cell 16 (1): 405–420. January 2005. doi:10.1091/mbc.E04-06-0516. PMID 15525674.
- ↑ 108.0 108.1 108.2 "TDP-43 is recruited to stress granules in conditions of oxidative insult". Journal of Neurochemistry 111 (4): 1051–1061. November 2009. doi:10.1111/j.1471-4159.2009.06383.x. PMID 19765185.
- ↑ 109.0 109.1 109.2 "C-Jun N-terminal kinase controls TDP-43 accumulation in stress granules induced by oxidative stress". Molecular Neurodegeneration 6: 57. August 2011. doi:10.1186/1750-1326-6-57. PMID 21819629.
- ↑ "HuD distribution changes in response to heat shock but not neurotrophic stimulation". The Journal of Histochemistry and Cytochemistry 54 (10): 1129–1138. October 2006. doi:10.1369/jhc.6A6979.2006. PMID 16801526.
- ↑ "Regulation of Human Endonuclease V Activity and Relocalization to Cytoplasmic Stress Granules". The Journal of Biological Chemistry 291 (41): 21786–21801. October 2016. doi:10.1074/jbc.M116.730911. PMID 27573237.
- ↑ 112.0 112.1 "The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response". BMC Cell Biology 9: 37. July 2008. doi:10.1186/1471-2121-9-37. PMID 18620564.
- ↑ 113.0 113.1 "FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations". Brain 134 (Pt 9): 2595–2609. September 2011. doi:10.1093/brain/awr201. PMID 21856723.
- ↑ "FAM98A is localized to stress granules and associates with multiple stress granule-localized proteins". Molecular and Cellular Biochemistry 451 (1–2): 107–115. January 2019. doi:10.1007/s11010-018-3397-6. PMID 29992460.
- ↑ 115.0 115.1 115.2 115.3 "Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression". Human Molecular Genetics 11 (24): 3007–3017. November 2002. doi:10.1093/hmg/11.24.3007. PMID 12417522.
- ↑ 116.0 116.1 "Oxidative stress reveals heterogeneity of FMRP granules in PC12 cell neurites". Brain Research 1112 (1): 56–64. September 2006. doi:10.1016/j.brainres.2006.07.026. PMID 16919243.
- ↑ 117.0 117.1 117.2 "Identification of the junctional plaque protein plakophilin 3 in cytoplasmic particles containing RNA-binding proteins and the recruitment of plakophilins 1 and 3 to stress granules". Molecular Biology of the Cell 17 (3): 1388–1398. March 2006. doi:10.1091/mbc.E05-08-0708. PMID 16407409.
- ↑ 118.0 118.1 118.2 118.3 "PKCα binds G3BP2 and regulates stress granule formation following cellular stress". PLOS ONE 7 (4): e35820. 2012. doi:10.1371/journal.pone.0035820. PMID 22536444. Bibcode: 2012PLoSO...735820K.
- ↑ "Both G3BP1 and G3BP2 contribute to stress granule formation". Genes to Cells 18 (2): 135–146. February 2013. doi:10.1111/gtc.12023. PMID 23279204.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs named:8 - ↑ "Cytoplasmic hGle1A regulates stress granules by modulation of translation". Molecular Biology of the Cell 26 (8): 1476–1490. April 2015. doi:10.1091/mbc.E14-11-1523. PMID 25694449.
- ↑ 122.00 122.01 122.02 122.03 122.04 122.05 122.06 122.07 122.08 122.09 122.10 122.11 122.12 122.13 122.14 122.15 122.16 122.17 122.18 122.19 "Stress Granule Assembly Disrupts Nucleocytoplasmic Transport". Cell 173 (4): 958–971.e17. May 2018. doi:10.1016/j.cell.2018.03.025. PMID 29628143.
- ↑ 123.0 123.1 "Regulation of stress granule dynamics by Grb7 and FAK signalling pathway". The EMBO Journal 27 (5): 715–726. March 2008. doi:10.1038/emboj.2008.19. PMID 18273060.
- ↑ 124.0 124.1 "Syk Is Recruited to Stress Granules and Promotes Their Clearance through Autophagy". The Journal of Biological Chemistry 290 (46): 27803–27815. November 2015. doi:10.1074/jbc.M115.642900. PMID 26429917.
- ↑ "Heat shock-induced accumulation of translation elongation and termination factors precedes assembly of stress granules in S. cerevisiae". PLOS ONE 8 (2): e57083. 2013. doi:10.1371/journal.pone.0057083. PMID 23451152. Bibcode: 2013PLoSO...857083G.
- ↑ "Evidence for the association of the human regulatory protein Ki-1/57 with the translational machinery". FEBS Letters 585 (16): 2556–2560. August 2011. doi:10.1016/j.febslet.2011.07.010. PMID 21771594.
- ↑ 127.0 127.1 127.2 Cite error: Invalid
<ref>tag; no text was provided for refs namedKwon_2007 - ↑ 128.0 128.1 "hnRNP A1 relocalization to the stress granules reflects a role in the stress response". Molecular and Cellular Biology 26 (15): 5744–5758. August 2006. doi:10.1128/MCB.00224-06. PMID 16847328.
- ↑ 129.0 129.1 "TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor". Molecular and Cellular Biology 31 (5): 1098–1108. March 2011. doi:10.1128/MCB.01279-10. PMID 21173160.
- ↑ "HuR-hnRNP interactions and the effect of cellular stress". Molecular and Cellular Biochemistry 372 (1–2): 137–147. January 2013. doi:10.1007/s11010-012-1454-0. PMID 22983828.
- ↑ "Molecular epidemiological study of familial amyotrophic lateral sclerosis in Japanese population by whole-exome sequencing and identification of novel HNRNPA1 mutation". Neurobiology of Aging 61: 255.e9–255.e16. January 2018. doi:10.1016/j.neurobiolaging.2017.08.030. PMID 29033165.
- ↑ 132.0 132.1 "TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1". Human Molecular Genetics 20 (7): 1400–1410. April 2011. doi:10.1093/hmg/ddr021. PMID 21257637.
- ↑ 133.0 133.1 "hnRNP K interacts with RNA binding motif protein 42 and functions in the maintenance of cellular ATP level during stress conditions". Genes to Cells 14 (2): 113–128. February 2009. doi:10.1111/j.1365-2443.2008.01256.x. PMID 19170760.
- ↑ "A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism". Molecular Cell 63 (5): 796–810. September 2016. doi:10.1016/j.molcel.2016.07.021. PMID 27570075.
- ↑ "The Co-Chaperone HspBP1 Is a Novel Component of Stress Granules that Regulates Their Formation". Cells 9 (4): 825. March 2020. doi:10.3390/cells9040825. PMID 32235396.
- ↑ "NF90 exerts antiviral activity through regulation of PKR phosphorylation and stress granules in infected cells". Journal of Immunology 192 (8): 3753–3764. April 2014. doi:10.4049/jimmunol.1302813. PMID 24623135.
- ↑ "Intracellular localization of human Ins(1,3,4,5,6)P5 2-kinase". The Biochemical Journal 408 (3): 335–345. December 2007. doi:10.1042/BJ20070382. PMID 17705785.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedTsai_2017 - ↑ "Stable formation of compositionally unique stress granules in virus-infected cells". Journal of Virology 84 (7): 3654–3665. April 2010. doi:10.1128/JVI.01320-09. PMID 20106928.
- ↑ "Sam68 relocalization into stress granules in response to oxidative stress through complexing with TIA-1". Experimental Cell Research 315 (19): 3381–3395. November 2009. doi:10.1016/j.yexcr.2009.07.011. PMID 19615357.
- ↑ "The nuclear protein Sam68 is recruited to the cytoplasmic stress granules during enterovirus 71 infection". Microbial Pathogenesis 96: 58–66. July 2016. doi:10.1016/j.micpath.2016.04.001. PMID 27057671.
- ↑ "Identification of FUSE-binding proteins as interacting partners of TIA proteins". Biochemical and Biophysical Research Communications 343 (1): 57–68. April 2006. doi:10.1016/j.bbrc.2006.02.112. PMID 16527256.
- ↑ 143.0 143.1 143.2 143.3 "Identification of Novel Stress Granule Components That Are Involved in Nuclear Transport". PLOS ONE 8 (6): e68356. 2013. doi:10.1371/journal.pone.0068356. PMID 23826389. Bibcode: 2013PLoSO...868356M.
- ↑ 144.0 144.1 "Identification of importin alpha1 as a novel constituent of RNA stress granules". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1803 (7): 865–871. July 2010. doi:10.1016/j.bbamcr.2010.03.020. PMID 20362631.
- ↑ "La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability". Molecular and Cellular Biology 31 (3): 542–556. February 2011. doi:10.1128/MCB.01162-10. PMID 21098120.
- ↑ 146.0 146.1 "Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules". RNA Biology 4 (1): 16–25. January 2007. doi:10.4161/rna.4.1.4364. PMID 17617744.
- ↑ 147.0 147.1 "The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci". RNA 8 (12): 1489–1501. December 2002. doi:10.1017/S1355838202021726. PMID 12515382.
- ↑ "RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules". RNA 12 (4): 547–554. April 2006. doi:10.1261/rna.2302706. PMID 16484376.
- ↑ 149.0 149.1 "Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP". The Journal of Cell Biology 181 (4): 639–653. May 2008. doi:10.1083/jcb.200708004. PMID 18490513.
- ↑ "Proteomic analysis reveals that MAEL, a component of nuage, interacts with stress granule proteins in cancer cells". Oncology Reports 31 (1): 342–350. January 2014. doi:10.3892/or.2013.2836. PMID 24189637.
- ↑ 151.0 151.1 151.2 "A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation". Molecular Biology of the Cell 21 (1): 117–130. January 2010. doi:10.1091/mbc.E09-06-0512. PMID 19910486.
- ↑ 152.0 152.1 "Interaction with 14-3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules". The Journal of Biological Chemistry 283 (46): 32131–32142. November 2008. doi:10.1074/jbc.M802927200. PMID 18779327.
- ↑ "Pivotal role of RNA-binding E3 ubiquitin ligase MEX3C in RIG-I-mediated antiviral innate immunity". Proceedings of the National Academy of Sciences of the United States of America 111 (15): 5646–5651. April 2014. doi:10.1073/pnas.1401674111. PMID 24706898. Bibcode: 2014PNAS..111.5646K.
- ↑ "Musashi-1 maintains blood-testis barrier structure during spermatogenesis and regulates stress granule formation upon heat stress". Molecular Biology of the Cell 26 (10): 1947–1956. May 2015. doi:10.1091/mbc.E14-11-1497. PMID 25717188.
- ↑ "MTHFSD and DDX58 are novel RNA-binding proteins abnormally regulated in amyotrophic lateral sclerosis". Brain 139 (Pt 1): 86–100. January 2016. doi:10.1093/brain/awv308. PMID 26525917.
- ↑ 156.0 156.1 156.2 156.3 156.4 156.5 "The mTOR-S6 kinase pathway promotes stress granule assembly". Cell Death and Differentiation 25 (10): 1766–1780. November 2018. doi:10.1038/s41418-018-0076-9. PMID 29523872.
- ↑ "An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response". Molecular Cell 25 (5): 765–778. March 2007. doi:10.1016/j.molcel.2007.01.025. PMID 17349961.
- ↑ 158.0 158.1 "Interaction and colocalization of HERMES/RBPMS with NonO, PSF, and G3BP1 in neuronal cytoplasmic RNP granules in mouse retinal line cells". Genes to Cells 20 (4): 257–266. April 2015. doi:10.1111/gtc.12224. PMID 25651939.
- ↑ "OASL1 Traps Viral RNAs in Stress Granules to Promote Antiviral Responses". Molecules and Cells 41 (3): 214–223. March 2018. doi:10.14348/molcells.2018.2293. PMID 29463066.
- ↑ "OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress". Molecular and Cellular Biology 30 (8): 2006–2016. April 2010. doi:10.1128/MCB.01350-09. PMID 20154146.
- ↑ "Inactivation by oxidation and recruitment into stress granules of hOGG1 but not APE1 in human cells exposed to sub-lethal concentrations of cadmium". Mutation Research 685 (1–2): 61–69. March 2010. doi:10.1016/j.mrfmmm.2009.09.013. PMID 19800894.
- ↑ "New roles for the de-ubiquitylating enzyme OTUD4 in an RNA-protein network and RNA granules". Journal of Cell Science 132 (12): jcs229252. June 2019. doi:10.1242/jcs.229252. PMID 31138677.
- ↑ 163.0 163.1 163.2 Cite error: Invalid
<ref>tag; no text was provided for refs namedKedersha_1999 - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedRalser_2005 - ↑ 165.0 165.1 165.2 165.3 165.4 165.5 "Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm". Molecular Cell 42 (4): 489–499. May 2011. doi:10.1016/j.molcel.2011.04.015. PMID 21596313.
- ↑ 166.0 166.1 "The Parkinson's Disease-Linked Protein DJ-1 Associates with Cytoplasmic mRNP Granules During Stress and Neurodegeneration". Molecular Neurobiology 56 (1): 61–77. January 2019. doi:10.1007/s12035-018-1084-y. PMID 29675578.
- ↑ "PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions". Scientific Reports 7 (1): 14035. October 2017. doi:10.1038/s41598-017-14156-8. PMID 29070863. Bibcode: 2017NatSR...714035C.
- ↑ "Pdcd4 Is Involved in the Formation of Stress Granule in Response to Oxidized Low-Density Lipoprotein or High-Fat Diet". PLOS ONE 11 (7): e0159568. 2016. doi:10.1371/journal.pone.0159568. PMID 27454120. Bibcode: 2016PLoSO..1159568B.
- ↑ 169.0 169.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedFigley_2014 - ↑ 170.0 170.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedHua_2004 - ↑ 171.0 171.1 171.2 "ZFAND1 Recruits p97 and the 26S Proteasome to Promote the Clearance of Arsenite-Induced Stress Granules" (in English). Molecular Cell 70 (5): 906–919.e7. June 2018. doi:10.1016/j.molcel.2018.04.021. PMID 29804830.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedDi_Salvio_2005 - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedDaigle_2016 - ↑ "Polysome-bound endonuclease PMR1 is targeted to stress granules via stress-specific binding to TIA-1". Molecular and Cellular Biology 26 (23): 8803–8813. December 2006. doi:10.1128/MCB.00090-06. PMID 16982678.
- ↑ 175.0 175.1 "Stress granules inhibit apoptosis by reducing reactive oxygen species production". Molecular and Cellular Biology 33 (4): 815–829. February 2013. doi:10.1128/MCB.00763-12. PMID 23230274.
- ↑ 176.0 176.1 176.2 "Stress Granules Contain Rbfox2 with Cell Cycle-related mRNAs". Scientific Reports 7 (1): 11211. September 2017. doi:10.1038/s41598-017-11651-w. PMID 28894257. Bibcode: 2017NatSR...711211P.
- ↑ 177.0 177.1 "Stress-dependent miR-980 regulation of Rbfox1/A2bp1 promotes ribonucleoprotein granule formation and cell survival". Nature Communications 9 (1): 312. January 2018. doi:10.1038/s41467-017-02757-w. PMID 29358748. Bibcode: 2018NatCo...9..312K.
- ↑ "Cell stress modulates the function of splicing regulatory protein RBM4 in translation control". Proceedings of the National Academy of Sciences of the United States of America 104 (7): 2235–2240. February 2007. doi:10.1073/pnas.0611015104. PMID 17284590. Bibcode: 2007PNAS..104.2235L.
- ↑ "Identification of the RNA recognition element of the RBPMS family of RNA-binding proteins and their transcriptome-wide mRNA targets". RNA 20 (7): 1090–1102. July 2014. doi:10.1261/rna.045005.114. PMID 24860013.
- ↑ 180.0 180.1 "The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs". The FEBS Journal 277 (9): 2109–2127. May 2010. doi:10.1111/j.1742-4658.2010.07628.x. PMID 20412057.
- ↑ "Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival". Molecular Cell 31 (5): 722–736. September 2008. doi:10.1016/j.molcel.2008.06.025. PMID 18775331.
- ↑ 182.0 182.1 "Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules". The Journal of Biological Chemistry 280 (52): 43131–43140. December 2005. doi:10.1074/jbc.M508374200. PMID 16221671.
- ↑ "Localization of SERBP1 in stress granules and nucleoli". The FEBS Journal 281 (1): 352–364. January 2014. doi:10.1111/febs.12606. PMID 24205981.
- ↑ "Stress granules counteract senescence by sequestration of PAI-1". EMBO Reports 19 (5): e44722. May 2018. doi:10.15252/embr.201744722. PMID 29592859.
- ↑ "The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals". Journal of Cell Science 126 (Pt 22): 5166–5177. November 2013. doi:10.1242/jcs.130708. PMID 24013546.
- ↑ 186.0 186.1 186.2 "A novel role for hSMG-1 in stress granule formation". Molecular and Cellular Biology 31 (22): 4417–4429. November 2011. doi:10.1128/MCB.05987-11. PMID 21911475.
- ↑ 187.0 187.1 187.2 "Survival motor neuron protein facilitates assembly of stress granules". FEBS Letters 572 (1–3): 69–74. August 2004. doi:10.1016/j.febslet.2004.07.010. PMID 15304326.
- ↑ "SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress". Cellular and Molecular Neurobiology 31 (4): 541–550. May 2011. doi:10.1007/s10571-011-9647-8. PMID 21234798.
- ↑ "Poly(A)(+) mRNA-binding protein Tudor-SN regulates stress granules aggregation dynamics". The FEBS Journal 282 (5): 874–890. March 2015. doi:10.1111/febs.13186. PMID 25559396.
- ↑ "Arsenite-activated JNK signaling enhances CPEB4-Vinexin interaction to facilitate stress granule assembly and cell survival". PLOS ONE 9 (9): e107961. 2014. doi:10.1371/journal.pone.0107961. PMID 25237887. Bibcode: 2014PLoSO...9j7961C.
- ↑ "SGNP: an essential Stress Granule/Nucleolar Protein potentially involved in 5.8s rRNA processing/transport". PLOS ONE 3 (11): e3716. 2008. doi:10.1371/journal.pone.0003716. PMID 19005571. Bibcode: 2008PLoSO...3.3716Z.
- ↑ "Direct binding of the Alu binding protein dimer SRP9/14 to 40S ribosomal subunits promotes stress granule formation and is regulated by Alu RNA". Nucleic Acids Research 42 (17): 11203–11217. 2014. doi:10.1093/nar/gku822. PMID 25200073.
- ↑ "The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression". The FEBS Journal 277 (11): 2496–2514. June 2010. doi:10.1111/j.1742-4658.2010.07664.x. PMID 20477871.
- ↑ "Poliovirus infection induces the co-localization of cellular protein SRp20 with TIA-1, a cytoplasmic stress granule protein". Virus Research 176 (1–2): 223–231. September 2013. doi:10.1016/j.virusres.2013.06.012. PMID 23830997.
- ↑ "Oxidative stress-inducible truncated serine/arginine-rich splicing factor 3 regulates interleukin-8 production in human colon cancer cells". American Journal of Physiology. Cell Physiology 306 (3): C250–C262. February 2014. doi:10.1152/ajpcell.00091.2013. PMID 24284797.
- ↑ "NEDDylation promotes stress granule assembly". Nature Communications 7: 12125. July 2016. doi:10.1038/ncomms12125. PMID 27381497. Bibcode: 2016NatCo...712125J.
- ↑ 197.0 197.1 "Calcium-responsive transactivator (CREST) protein shares a set of structural and functional traits with other proteins associated with amyotrophic lateral sclerosis". Molecular Neurodegeneration 10: 20. April 2015. doi:10.1186/s13024-015-0014-y. PMID 25888396.
- ↑ "Mammalian Staufen 1 is recruited to stress granules and impairs their assembly". Journal of Cell Science 122 (Pt 4): 563–573. February 2009. doi:10.1242/jcs.038208. PMID 19193871.
- ↑ "Human hnRNP Q re-localizes to cytoplasmic granules upon PMA, thapsigargin, arsenite and heat-shock treatments". Experimental Cell Research 315 (6): 968–980. April 2009. doi:10.1016/j.yexcr.2009.01.012. PMID 19331829.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedBlechingberg_2012 - ↑ "Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue". PLOS ONE 5 (10): e13250. October 2010. doi:10.1371/journal.pone.0013250. PMID 20948999. Bibcode: 2010PLoSO...513250L.
- ↑ "Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery". Journal of Proteome Research 9 (2): 1104–1120. February 2010. doi:10.1021/pr901076y. PMID 20020773.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedLi_2015 - ↑ 204.0 204.1 "TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics". Neuron 95 (4): 808–816.e9. August 2017. doi:10.1016/j.neuron.2017.07.025. PMID 28817800.
- ↑ "TDP-43 regulation of stress granule dynamics in neurodegenerative disease-relevant cell types". Scientific Reports 8 (1): 7551. May 2018. doi:10.1038/s41598-018-25767-0. PMID 29765078. Bibcode: 2018NatSR...8.7551K.
- ↑ "Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP". Human Molecular Genetics 17 (20): 3236–3246. October 2008. doi:10.1093/hmg/ddn219. PMID 18664458.
- ↑ 207.0 207.1 "Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders". Nature Neuroscience 16 (9): 1228–1237. September 2013. doi:10.1038/nn.3484. PMID 23912948.
- ↑ 208.0 208.1 "Arginine methylation of USP9X promotes its interaction with TDRD3 and its anti-apoptotic activities in breast cancer cells". Cell Discovery 3: 16048. 2017. doi:10.1038/celldisc.2016.48. PMID 28101374.
- ↑ "Cytoplasmic TERT Associates to RNA Granules in Fully Mature Neurons: Role in the Translational Control of the Cell Cycle Inhibitor p15INK4B". PLOS ONE 8 (6): e66602. 2013. doi:10.1371/journal.pone.0066602. PMID 23825548. Bibcode: 2013PLoSO...866602I.
- ↑ "TIA1 variant drives myodegeneration in multisystem proteinopathy with SQSTM1 mutations". The Journal of Clinical Investigation 128 (3): 1164–1177. March 2018. doi:10.1172/JCI97103. PMID 29457785.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedBakkar_2005 - ↑ "A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay". Nucleic Acids Research 37 (19): 6600–6612. October 2009. doi:10.1093/nar/gkp717. PMID 19729507.
- ↑ "Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains". Cell 173 (3): 677–692.e20. April 2018. doi:10.1016/j.cell.2018.03.002. PMID 29677512.
- ↑ "Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization". Nucleic Acids Research 46 (6): 3061–3074. April 2018. doi:10.1093/nar/gky103. PMID 29471495.
- ↑ "RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage". Genes & Development 24 (15): 1590–1595. August 2010. doi:10.1101/gad.586710. PMID 20679393.
- ↑ "UBAP2L arginine methylation by PRMT1 modulates stress granule assembly". Cell Death and Differentiation 27 (1): 227–241. January 2020. doi:10.1038/s41418-019-0350-5. PMID 31114027.
- ↑ "UBAP2L Forms Distinct Cores that Act in Nucleating Stress Granules Upstream of G3BP1". Current Biology 30 (4): 698–707.e6. February 2020. doi:10.1016/j.cub.2019.12.020. PMID 31956030.
- ↑ "Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions". Molecular Cell 69 (6): 965–978.e6. March 2018. doi:10.1016/j.molcel.2018.02.004. PMID 29526694.
- ↑ 219.0 219.1 219.2 "ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97" (in English). Molecular Cell 74 (4): 742–757.e8. May 2019. doi:10.1016/j.molcel.2019.03.027. PMID 30979586.
- ↑ 220.0 220.1 "Deubiquitylases USP5 and USP13 are recruited to and regulate heat-induced stress granules through their deubiquitylating activities". Journal of Cell Science 131 (8): jcs210856. April 2018. doi:10.1242/jcs.210856. PMID 29567855.
- ↑ "Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function". Cell 153 (7): 1461–1474. June 2013. doi:10.1016/j.cell.2013.05.037. PMID 23791177.
- ↑ "YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1". The Journal of Cell Biology 208 (7): 913–929. March 2015. doi:10.1083/jcb.201411047. PMID 25800057.
- ↑ 223.0 223.1 223.2 223.3 "m6A enhances the phase separation potential of mRNA". Nature 571 (7765): 424–428. July 2019. doi:10.1038/s41586-019-1374-1. PMID 31292544.
- ↑ 224.0 224.1 224.2 224.3 "m6A-binding YTHDF proteins promote stress granule formation". Nature Chemical Biology 16 (9): 955–963. September 2020. doi:10.1038/s41589-020-0524-y. PMID 32451507.
- ↑ "ZBP1 regulates mRNA stability during cellular stress". The Journal of Cell Biology 175 (4): 527–534. November 2006. doi:10.1083/jcb.200608071. PMID 17101699.
- ↑ "ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains". Nucleic Acids Research 34 (18): 5007–5020. 2006. doi:10.1093/nar/gkl575. PMID 16990255.
- ↑ "MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay". The EMBO Journal 23 (6): 1313–1324. March 2004. doi:10.1038/sj.emboj.7600163. PMID 15014438.
- ↑ "Protor-2 interacts with tristetraprolin to regulate mRNA stability during stress". Cellular Signalling 24 (1): 309–315. January 2012. doi:10.1016/j.cellsig.2011.09.015. PMID 21964062.
- ↑ "Recruitment of mRNA-destabilizing protein TIS11 to stress granules is mediated by its zinc finger domain". Experimental Cell Research 303 (2): 287–299. February 2005. doi:10.1016/j.yexcr.2004.09.031. PMID 15652343.
Further reading
- "RNA granules". The Journal of Cell Biology 172 (6): 803–808. March 2006. doi:10.1083/jcb.200512082. PMID 16520386.
- "Stress granules: sites of mRNA triage that regulate mRNA stability and translatability". Biochemical Society Transactions 30 (Pt 6): 963–969. November 2002. doi:10.1042/BST0300963. PMID 12440955.
— molecular details of stress granule assembly & function - "Nuclear stress granules: the awakening of a sleeping beauty?". The Journal of Cell Biology 164 (1): 15–17. January 2004. doi:10.1083/jcb.200311102. PMID 14709538.
External links
Laboratories:
- Anderson lab, post-transcriptional control & inflammatory response
- Zhou lab, neurodegenerative disease
- Morimoto lab, transcriptional regulation & heat shock
- Boccaccio lab, Staufen & stress granules
 |