Biology:Haplogroup E-M2
| Haplogroup E-M2 (former E3a / E1b1a) | |
|---|---|
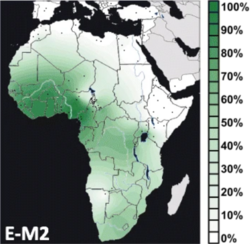 Interpolated frequency distribution.[1] | |
| Possible time of origin | 39,200 years BP[2] |
| Coalescence age | 16,300 years BP[2] |
| Possible place of origin | West Africa[3][4] or Central Africa[3][4] |
| Ancestor | E-V38 |
| Descendants | E-Z5994, E-V43 |
| Defining mutations | M2, DYS271/SY81, M291, P1/PN1, P189.1, P293.1 |
Haplogroup E-M2, also known as E1b1a1-M2, is a human Y-chromosome DNA haplogroup. E-M2 is primarily distributed within sub-Saharan Africa. More specifically, E-M2 is the predominant subclade in West Africa, Central Africa, Southern Africa, and the region of the African Great Lakes; it also occurs at low to moderate frequencies in North Africa, and at low frequencies in the Middle East. E-M2 has several subclades, but many of these subhaplogroups are included in either E-L485 or E-U175. E-M2 is especially common among indigenous Africans who speak Niger-Congo languages, and was spread to Southern Africa and East Africa through the Bantu expansion.
Origins
The discovery of two SNPs (V38 and V100) by Trombetta et al. (2011) significantly redefined the E-V38 phylogenetic tree. This led the authors to suggest that E-V38 may have originated in East Africa. E-V38 joins the West African-affiliated E-M2 and the Northeast African-affiliated E-M329 with an earlier common ancestor who, like E-P2, may have also originated in East Africa.[5] The downstream SNP E-M180 may have originated in the humid south-central Saharan savanna/grassland of North Africa between 14,000 BP and 10,000 BP.[6][7][8][9] According to Wood et al. (2005) and Rosa et al. (2007), such population movements changed the pre-existing population Y chromosomal diversity in Central, Southern, and Southeastern Africa, replacing the previous haplogroup frequencies in these areas with the now dominant E1b1a1 lineages. Traces of earlier inhabitants, however, can be observed today in these regions via the presence of the Y DNA haplogroups A1a, A1b, A2, A3, and B-M60 that are common in certain populations, such as the Mbuti and Khoisan.[10][11][12] Shriner et al. (2018) similarly suggests that haplogroup E1b1a-V38 migrated across the Green Sahara from east to west around 19,000 years ago, where E1b1a1-M2 may have subsequently originated in West Africa or Central Africa. Shriner et al. (2018) also traces this migration via sickle cell mutation, which likely originated during the Green Sahara period.[4]
Ancient DNA
Within Africa
Botswana
At Xaro, in Botswana, there were two individuals, dated to the Early Iron Age (1400 BP); one carried haplogroups E1b1a1a1c1a and L3e1a2, and another carried haplogroups E1b1b1b2b (E-M293, E-CTS10880) and L0k1a2.[13][14]
At Taukome, in Botswana, an individual, dated to the Early Iron Age (1100 BP), carried haplogroups E1b1a1 (E-M2, E-Z1123) and L0d3b1.[13][14]
Democratic Republic of Congo
At Kindoki, in the Democratic Republic of Congo, there were three individuals, dated to the protohistoric period (230 BP, 150 BP, 230 BP); one carried haplogroups E1b1a1a1d1a2 (E-CTS99, E-CTS99) and L1c3a1b, another carried haplogroup E (E-M96, E-PF1620), and the last carried haplogroups R1b1 (R-P25 1, R-M415) and L0a1b1a1.[13][14]
Egypt
Hawass et al. (2012) determined that the ancient Egyptian mummy of an unknown man buried with Ramesses was, because of the proven genetic relationship and a mummification process that suggested punishment, a good candidate for the pharaoh's son, Pentaweret, who was the only son to revolt against his father.[15] It was impossible to determine his cause of death.[15] Using Whit Athey's haplogroup predictor based on Y-STR values, both mummies were predicted to share the Y chromosomal haplogroup E1b1a1-M2 and 50% of their genetic material, which pointed to a father-son relationship.[15] Gad et al. (2021) indicates that Ramesses III and Unknown Man E, possibly Pentawere, carried haplogroup E1b1a.[16]
Kenya
At Deloraine Farm, in Nakuru County, Kenya, an iron metallurgist of the Iron Age carried haplogroups E1b1a1a1a1a/E-M58 and L5b1.[17][18]
At Lamu, Pate Island, Faza, in Kenya, an individual, dated between 1500 CE and 1700 CE, carried haplogroups E1b1a1a1a2a1a and L3e3a.[19]
At Taita Taveta, Makwasinyi, in Kenya, an individual, dated between 1650 CE and 1950 CE, carried haplogroups E1b1a1a1a2a1a and L4b2a.[19]
At Taita Taveta, Makwasinyi, in Kenya, an individual, dated between 1650 CE and 1950 CE, carried haplogroups E1b1a1a1a2a1a3b1d1c and L1c3b1a.[19]
At Taita Taveta, Makwasinyi, in Kenya, an individual, dated between 1650 CE and 1950 CE, carried haplogroups E1b1a1a1a2a1a and L2a1+143.[19]
At Taita Taveta, Makwasinyi, in Kenya, an individual, dated between 1667 cal CE and 1843 cal CE, carried haplogroups E1b1a1a1a2a1a3b1d1c and L2a1+143.[19]
At Taita Taveta, Makwasinyi, in Kenya, an individual, dated between 1709 cal CE and 1927 cal CE, carried haplogroups E1b1a1a1a2a1a3a1d~ and L3a2.[19]
Tanzania
At Songo Mnara, in Tanzania, an individual, dated between 1418 cal CE and 1450 cal CE, carried haplogroups E1b1a1~ and L3e2b.[19]
At Lindi, in Tanzania, an individual, dated between 1511 cal CE and 1664 cal CE, carried haplogroups E1b1a1a1a2a1a3a1d~ and L0a1a2.[19]
Outside of Africa
Mexico
At a San Jose de los Naturales Royal Hospital burial site, in Mexico City, Mexico, three enslaved West Africans of West African and Southern African ancestry, dated between 1453 CE and 1626 CE, 1450 CE and 1620 CE, and 1436 CE and 1472 CE, were found; one carried haplogroups E1b1a1a1c1b/E-M263.2 and L1b2a, another carried haplogroups E1b1a1a1d1/E-P278.1/E-M425 and L3d1a1a, and the last carried haplogroups E1b1a1a1c1a1c/E-CTS8030 and L3e1a1a.[20] Human leukocyte antigen alleles further confirm that the individuals were of Sub-Saharan African origin.[21]
Portugal
At Cabeço da Amoreira, in Portugal, an enslaved West African man, who may have been from the Senegambian coastal region of Gambia, Mauritania, or Senegal, and carried haplogroups E1b1a and L3b1a, was buried among shell middens between the 16th century CE and the 18th century CE.[22]
Saint Helena
In Saint Helena, 20 freed Africans,[23][24] who were dated to the 19th century CE,[23] were also of western Central African[23][25][26] (e.g., Bantu peoples of Gabon and Angola) ancestry.[23] One female individual carried haplogroup L1b1a10b.[27] One female individual carried haplogroup L2a1f.[27] One female individual carried haplogroup L2a1a3c.[27] One male individual carried haplogroups E1b1a1a1a2a1a3b1d and L1c3a.[27] One male individual carried haplogroups E1b1a1a1a1c1a1a and L0a1b2a.[27] One male individual carried haplogroups E1b1a1a1a2a1a3b1a2a2 and L0a1e.[27] One male individual carried haplogroups E1b1a1a1a2a1a3b1 and L2a1f1.[27] One male individual carried haplogroups E1b1a1 and L3.[27] One male individual carried haplogroups E1b1a1a1a2a1a3b1d and L3e1e.[27] One male individual carried haplogroups E1b1a1a1a2a1a3a1d and L3e3b2.[27] One male individual carried haplogroups E1b1a1a1a1c1a1a3 and L3e1a3a.[27] One male individual carried haplogroups E1b1a1a1a2a1a3b1a2a2 and L2b1a.[27] One male individual carried haplogroups E1b1a1a1a2a1a3b1 and L3f1b1a.[27] One male individual carried haplogroups E1b1a1a1a2a1a3b1d1c1a and L3d3a1.[27] One male individual carried haplogroups B2a1a1a1 and L3e2b1.[27] One male individual carried haplogroups E1b1a1a1a2a1a3b1d1c1a and L2a1f.[27] One male individual carried haplogroups E1b1a1a1a1c1a1a3a1c1 and L3e1d1a.[27] One male individual carried haplogroups E1b1a1a1a2a1a3a1d and L1b1a10.[27] One male individual carried haplogroups E1b1a1a1a1c1a1a3a1c and L2a1f1.[27] One male individual carried haplogroups E1b1a1a1a1c1a1 and L2b1a.[27] An enslaved African American man and woman, from the 18th century CE Anson Street burial site in Charleston, South Carolina, who carried haplogroup L3e1e, shared this haplogroup with freed Africans in Saint Helena.[28] Based on those who were present among enlaved Africans, the ratio of males-to-females supports the conclusion of there being a strong selection bias for males in the latter period of the Trans-Atlantic Slave Trade.[23][29][30] Consequently, due to this study on the freed Africans of Saint Helena, among other studies, greater genetic insights have been made into the Trans-Atlantic Slave Trade and its effects on the demographics of Africa.[31]
Spain
In Granada, a Muslim (Moor) of the Cordoba Caliphate,[32] who was of haplogroups E1b1a1 and H1+16189,[33][34] as well as estimated to date between 900 CE and 1000 CE, and a Morisco,[32] who was of haplogroup L2e1,[33][34] as well as estimated to date between 1500 CE and 1600 CE, were both found to be of West African (i.e., Gambian) and Iberian descent.[32]
United States of America
At Avery’s Rest, in Chesapeake, Delaware, 3 out of 11 individuals were African Americans, who were dated between 1675 CE and 1725 CE; one was of West African ancestry and carried haplogroups E1b1a-CTS2447 and L3e3b, another was of western Central African Bantu-speaking ancestry and carried E1b1a-Z5974 and L0a1a2, and another was of West African and East African ancestry and carried E1b1a-Z5974 and L3d2.[35]
At Catoctin Furnace African American Cemetery, in Catoctin Furnace, Maryland, there were 27 African Americans found who were dated between 1774 CE and 1850 CE.[36][37] One male individual, who was of 98.14% Sub-Saharan African ancestry, carried haplogroups E1b1a1a1a1c2c and L2a1+143+@16309.[38] One male individual, who was of 83.73% Sub-Saharan African and 7.74% European ancestry, carried haplogroups E1b1a1a1a1c1b1 and L3e2a1b1.[38] One male individual, who was of 84.94% Sub-Saharan African and 9.45% European ancestry, carried haplogroups E1b1a1a1a2a1a and L2a1+143+16189 (16192)+@16309.[38] One male individual, who was of 87.83% Sub-Saharan African and 8.23% European ancestry, carried haplogroups E1b1a1a1a1c1a1a3a1d1 and L3d1b3.[38] One male individual, who was of 98.14% Sub-Saharan African ancestry, carried haplogroups E1b1a1a1a1a and L3e2a1b1.[38] One male individual, who was of 93.87% Sub-Saharan African and 2.58% European ancestry, carried haplogroups E1b1a1a1 and L3e1.[38] One male individual, who was of 98.70% Sub-Saharan African ancestry, carried haplogroups E1b1a1a1a1c1b2a and L2a1a1.[38] One male individual, who was of 97.01% Sub-Saharan African ancestry, carried haplogroups E1b1a1a1a1c1a1 and L3e2a1b1.[38] One male individual, who was of 82.31% Sub-Saharan African and 10.24% European ancestry, carried haplogroups E1b1a1a1a1c1b and L3e2a1b1.[38] One male individual, who was of 91.82% Sub-Saharan African and 5.31% European ancestry, carried haplogroups E1b1a1a1a1c1a1 and L3e2.[38] One male individual, who was of 81.18% Sub-Saharan African and 14.86% European ancestry, carried haplogroups E1b1a1~ and L2c.[38]
At an Anson Street burial site, in Charleston, South Carolina, there were 18 African Americans found who were dated to the 18th century CE.[39] Banza was of western Central African ancestry and carried haplogroups E1b1a-CTS668 and L3e3b1.[39] Lima was of West African ancestry and carried haplogroups E1b1a-M4671 and L3b3.[39] Kuto was of western Central African ancestry and carried haplogroups E1b1a-CTS2198 and L2a1a2.[39] Anika was of Sub-Saharan African ancestry and carried haplogroups E1b1a-CTS6126 and L2b1.[39] Nana was of West African ancestry and carried haplogroup L2b3a.[39] Zimbu was of western Central African ancestry and carried haplogroups E1b1a-CTS5497 and L3e1e.[39] Wuta was of Sub-Saharan African ancestry and carried haplogroups E1b1a-CTS7305 and L3e2b+152.[39] Daba was of West African ancestry and carried haplogroups E1b1a-M4273 and L2c.[39] Fumu was of Sub-Saharan African ancestry and carried haplogroups B2a1a-Y12201 and L3e2b+152.[39] Lisa was of West African ancestry and carried haplogroups E1b1a-Z6020 and H100.[39] Ganda was of West African ancestry and carried haplogroups E1b1a-CTS5612 and L1c1c.[39] Coosaw was of West African and Native American ancestry and carried haplogroups E2b1a-CTS2400 and A2.[39] Kidzera was of western Central African ancestry and carried haplogroup L2a1a2c.[39] Pita was of Sub-Saharan African ancestry and carried haplogroups E1b1a-M4287 and L3e2b.[39] Tima was of western Central African ancestry and carried haplogroup L3e1e.[39] Jode was of Sub-Saharan African ancestry and carried haplogroups E1b1a-CTS4975 and L2a1a2c.[39] Ajana was of western Central African ancestry and carried haplogroup L2a1I.[39] Isi was of western Central African ancestry and carried haplogroup L3e2a.[39]
Medical DNA
Sickle Cell
Amid the Green Sahara, the mutation for sickle cell originated in the Sahara[40] or in the northwest forest region of western Central Africa (e.g., Cameroon)[40][41] by at least 7,300 years ago,[40][41] though possibly as early as 22,000 years ago.[42][41] The ancestral sickle cell haplotype to modern haplotypes (e.g., Cameroon/Central African Republic and Benin/Senegal haplotypes) may have first arose in the ancestors of modern West Africans, bearing haplogroups E1b1a1-L485 and E1b1a1-U175 or their ancestral haplogroup E1b1a1-M4732.[40] West Africans (e.g., Yoruba and Esan of Nigeria), bearing the Benin sickle cell haplotype, may have migrated through the northeastern region of Africa into the western region of Arabia.[40] West Africans (e.g., Mende of Sierra Leone), bearing the Senegal sickle cell haplotype,[43][40] may have migrated into Mauritania (77% modern rate of occurrence) and Senegal (100%); they may also have migrated across the Sahara, into North Africa, and from North Africa, into Southern Europe, Turkey, and a region near northern Iraq and southern Turkey.[43] Some may have migrated into and introduced the Senegal and Benin sickle cell haplotypes into Basra, Iraq, where both occur equally.[43] West Africans, bearing the Benin sickle cell haplotype, may have migrated into the northern region of Iraq (69.5%), Jordan (80%), Lebanon (73%), Oman (52.1%), and Egypt (80.8%).[43]
Distribution
E-M2's frequency and diversity are highest in West Africa. Within Africa, E-M2 displays a west-to-east as well as a south-to-north clinal distribution. In other words, the frequency of the haplogroup decreases as one moves from western and southern Africa toward the eastern and northern parts of Africa.[44]
| Population group | frequency | References |
|---|---|---|
| Bamileke | 96%-100% | [44][45] |
| Ewe | 97% | [11] |
| Ga | 97% | [11] |
| Hutu | 94.2% | [44] |
| Yoruba | 93.1% | [46] |
| Tutsi | 32%-48% | [44] |
| Fante | 84% | [11] |
| Mandinka | 79%–87% | [10][11] |
| Ovambo | 82% | [11] |
| Senegalese | 81% | [47] |
| Ganda | 77% | [11] |
| Bijagós | 76% | [10] |
| Balanta | 73% | [10] |
| Fula | 73% | [10] |
| Kikuyu | 73% | [11] |
| Herero | 71% | [11] |
| Nalú | 71% | [10] |
Populations in Northwest Africa, central Eastern Africa and Madagascar have tested at more moderate frequencies.
| Population group | frequency | References |
|---|---|---|
| Tuareg from Tânout, Niger | 44.4% (8/18 subjects) | [48] |
| Comorian Shirazi | 41% | [49] |
| Tuareg from Gorom-Gorom, Burkina Faso | 16.6% (3/18) | [48] |
| Tuareg from Gossi, Mali | 9.1% (1/9) | [48] |
| Cape Verdeans | 15.9% (32/201) | [50] |
| Maasai | 15.4% (4/26) | [11] |
| Luo | 66% (6/9) | [11] |
| Iraqw | 11.11% (1/9) | [11] |
| Comoros | 23.46% (69/294) | [49] |
| Merina people (also called Highlanders) | 44% (4/9) | [51] |
| Antandroy | 69.6% (32/46) | [51] |
| Antanosy | 48.9% (23/47) | [51] |
| Antaisaka | 37.5% (3/8) | [51] |
E-M2 is found at low to moderate frequencies in North Africa, and Northeast Africa. Some of the lineages found in these areas are possibly due to the Bantu expansion or other migrations.[44][52] However, the discovery in 2011 of the E-M2 marker that predates E-M2 has led Trombetta et al. to suggest that E-M2 may have originated in East Africa.[5] In Eritrea and most of Ethiopia (excluding the Anuak), E-V38 is usually found in the form of E-M329, which is autochthonous, while E-M2 generally indicates Bantu migratory origins.[53][54][55]
| Population group | frequency | References |
|---|---|---|
| Tuareg from Al Awaynat and Tahala, Libya | 46.5% (20/43)[lower-alpha 1] | [56] |
| Oran, Algeria | 8.6% (8/93) | [57] |
| Berbers, southern and north-central Morocco | 9.5% (6/63) 5.8% (4/69) | [58][lower-alpha 2][59] |
| Moroccan Arabs | 6.8% (3/44) 1.9% (1/54) | [58][59] |
| Saharawis | 3.5% (1/29) | [58] |
| Egyptians | 1.4% (2/147), 0% (0/73), 8.33% (3/36) | [44][60][61] |
| Tunisians | 1.4% (2/148) | [61] |
| Sudanese (may include Hausa migrants) | 0.9% (4/445) | [62] |
| Somalia nationals (may include Bantu minorities) | 1.5% (3/201) | [52] |
Outside of Africa, E-M2 has been found at low frequencies. The clade has been found at low frequencies in West Asia. A few isolated occurrences of E-M2 have also been observed among populations in Southern Europe, such as Croatia, Malta, Spain and Portugal.[63][64][65][66]
| Population group | frequency | References |
|---|---|---|
| Bahrain | 8.6% (46/562) | |
| Saudi Arabians | 6.6% (11/157) | |
| Omanis | 6.6% (8/121) | [44] |
| Emiratis | 5.5% (9/164) | [69] |
| Yemenis | 4.8% (3/62) | [69] |
| Cypriots | 3.2% (2/62) | [66] |
| Southern Iranians | 1.7% (2/117) | [70] |
| Jordanians | 1.4% (2/139) | [71] |
| Sri Lanka | 1.4% (9/638) | [72] |
| Aeolian Islands, Italy | 1.2% (1/81) | [73] |
The Trans-Atlantic slave trade brought people to North America, Central America and South America including the Caribbean. Consequently, the haplogroup is often observed in the United States populations in men who self-identify as African Americans.[74] It has also been observed in a number of populations in Mexico, the Caribbean, Central America, and South America among people of African descent.
| Population group | frequency | References |
|---|---|---|
| Americans | 7.7–7.9%[lower-alpha 3] | [74] |
| Cubans | 9.8% (13/132) | [75] |
| Puerto Ricans | 19.23% (5/26) | [76] |
| Nicaraguans | 5.5% (9/165) | [77] |
| Several populations of Colombians | 6.18% (69/1116) | [78] |
| Alagoas, Brazil | 4.45% (11/247) | [79] |
| Bahia, Brazil | 19% (19/100) | [80] |
| Bahamians | 58.63% (251/428) | [81] |
Subclades
E1b1a1
E1b1a1 is defined by markers DYS271/M2/SY81, M291, P1/PN1, P189, P293, V43, and V95. Whilst E1b1a reaches its highest frequency of 81% in Senegal, only 1 of the 139 Senegalese that were tested showed M191/P86.[47] In other words, as one moves to West Africa from western Central Africa, the less subclade E1b1a1f is found. Cruciani et al. (2002) states: "A possible explanation might be that haplotype 24 chromosomes [E-M2*] were already present across the Sudanese belt when the M191 mutation, which defines haplotype 22, arose in central western Africa. Only then would a later demic expansion have brought haplotype 22 chromosomes from central western to western Africa, giving rise to the opposite clinal distributions of haplotypes 22 and 24."[45]
E1b1a1a1
E1b1a1a1 is commonly defined by M180/P88. The basal subclade is quite regularly observed in M2+ samples.
E1b1a1a1a
E1b1a1a1a is defined by marker M58. 5% (2/37) of the town Singa-Rimaïbé, Burkina Faso tested positive for E-M58.[45] 15% (10/69) of Hutus in Rwanda tested positive for M58.[44] Three South Africans tested positive for this marker.[12] One Carioca from Rio de Janeiro, Brazil tested positive for the M58 SNP.[82] The place of origin and age is unreported.
E1b1a1a1b
E1b1a1a1b is defined by M116.2, a private marker. A single carrier was found in Mali.[12][lower-alpha 4]
E1b1a1a1c
E1b1a1a1c is defined by private marker M149. This marker was found in a single South African.[12]
E1b1a1a1d
E1b1a1a1d is defined by a private marker M155. It is known from a single carrier in Mali.[12]
E1b1a1a1e
E1b1a1a1e is defined by markers M10, M66, M156 and M195. Wairak people in Tanzania tested 4.6% (2/43) positive for E-M10.[44] E-M10 was found in a single person of the Lissongo group in the Central African Republic and two members in a "Mixed" population from the Adamawa region.[12]
E1b1a1a1f
E1b1a1a1f is defined by L485. The basal node E-L485* appears to be somewhat uncommon but has not been sufficiently tested in large populations. The ancestral L485 SNP (along with several of its subclades) was very recently discovered. Some of these SNPs have little or no published population data and/or have yet to receive nomenclature recognition by the YCC.
- E1b1a1a1f1 is defined by marker L514. This SNP is currently without population study data outside of the 1000 Genomes Project.
- E1b1a1a1f1a (YCC E1b1a7) is defined by marker M191/P86. Filippo et al. (2011) studied a number of African populations that were E-M2 positive and found the basal E-M191/P86 (without E-P252/U174) in a population of Gur speakers in Burkina Faso.[83] Montano et al. (2011) found similar sparse distribution of E-M191* in Nigeria, Gabon, Cameroon and Congo.[9] M191/P86 positive samples occurred in tested populations of Annang (38.3%), Ibibio (45.6%), Efik (45%), and Igbo (54.3%) living in Nigeria, West Africa.[84] E-M191/P86 appears in varying frequencies in Central and Southern Africa but almost all are also positive for P252/U174. Bantu-speaking South Africans (89/343) tested 25.9% positive and Khoe-San speaking South Africans tested 7.7% (14/183) positive for this SNP.[85] It also appears commonly in Africans living in the Americas. A population in Rio de Janeiro, Brazil tested 9.2% (12/130) positive.[82] 34.9% (29/83) of African American men tested positive for M191.[74]
- Veeramah et al. (2010) studies of the recombining portions of M191 positive Y chromosomes suggest that this lineage has "diffusely spread with multiple high frequency haplotypes implying a longer evolutionary period since this haplogroup arose".[84] The subclade E1b1a1a1f1a appears to express opposite clinal distributions to E1b1a1* in the West African Savanna region. Haplogroup E1b1a1a1f1a (E-M191) has a frequency of 23% in Cameroon (where it represents 42% of haplotypes carrying the DYS271 mutation or E-M2), 13% in Burkina Faso (16% of haplotypes carrying the M2/DYS271 mutation) and only 1% in Senegal.[47] Similarly, while E1b1a reaches its highest frequency of 81% in Senegal, only 1 of the 139 Senegalese that were tested showed M191/P86.[47] In other words, as one moves to West Africa from western Central Africa, the less subclade E1b1a1f is found. "A possible explanation might be that haplotype 24 chromosomes [E-M2*] were already present across the Sudanese belt when the M191 mutation, which defines haplotype 22, arose in central western Africa. Only then would a later demic expansion have brought haplotype 22 chromosomes from central western to western Africa, giving rise to the opposite clinal distributions of haplotypes 22 and 24."[45]
- E1b1a1a1f1a1 (YCC E1b1a7a) is defined by P252/U174. It appears to be the most common subclade of E-L485. It is believed to have originated near western Central Africa. It is rarely found in the most western portions of West Africa. Montano et al. (2011) found this subclade very prevalent in Nigeria and Gabon.[9] Filippo et al. (2011) estimated a tMRCA of ~4.2 kya from sample of Yoruba population positive for the SNP.[83]
- E1b1a1a1f1a1b (YCC E1b1a7a2) is defined by P115. This subclade has only been observed amongst Fang people of Central Africa.[9]
- E1b1a1a1f1a1c (YCC E1b1a7a3) is defined by P116. Montano et al. (2011) observed this SNP only in Gabon and a Bassa population from Cameroon.[9]
- E1b1a1a1f1a1d is defined by Z1704. This subclade has been observed across Africa. The 1000 Genomes Project Consortium found this SNP in Yoruba Nigerian, three Kenyan Luhyas and one African descent Puerto Rican.[86]
- E1b1a1a1f1b is defined by markers L515, L516, L517, and M263.2. This subclade was found by the researchers of Y-Chromosome Genome Comparison Project using data from the commercial bioinformatics company 23andMe.[87]
E1b1a1a1g
E1b1a1a1g (YCC E1b1a8) is defined by marker U175. The basal E-U175* is extremely rare. Montano et al. (2011) only found one out of 505 tested African subjects who was U175 positive but negative for U209.[9] Brucato et al. found similarly low frequencies of basal E-U175* in subjects in the Ivory Coast and Benin. Veeramah et al. (2010) found U175 in tested Annang (45.3%), Ibibio (37%), Efik (33.3%), and Igbo (25.3%) but did not test for U209.[84]
The supposed "Bantu haplotype" found in E-U175 carriers is "present at appreciable frequencies in other Niger–Congo languages speaking peoples as far west as Guinea-Bissau".[84] This is the modal haplotype of STR markers that is common in carriers of E-U175.[lower-alpha 5]
| E-U175 haplotype | DYS19 | DYS388 | DYS390 | DYS391 | DYS392 | DYS393 | |
| 15 | 12 | 21 | 10 | 11 | 13 | ||
E1b1a1a1g has several subclades.
- E1b1a1a1g1 (YCC E1b1a8a) is defined by U209. It is the most prominent subclade of U175. This subclade has very high frequencies of over fifty percentages in Cameroonian populations of Bassa and Bakaka, possibly indicating place of origin. However, E-U209 is widely found at lower frequencies in West and Central African countries surrounding Cameroon and Gabon.[9] Brucato et al. (2010) found the SNP in a populations of Ahizi (in Ivory Coast) 38.8% (19/49), Yacouba (Ivory Coast) 27.5% (11/40), and Beninese 6.5% (5/77) respectively.[88]
- E1b1a1a1g1a (YCC E1b1a8a1) is defined by U290. The Montano et al. (2011) study of U290 showed a lower frequency in Nigeria (11.7%) and western Central Africa than basal node U209. The highest population frequency rate in that study was 57.7% (15/26) in Ewondo in Cameroon.[9] 32.5% (27/83) of African American men tested by Sims et al. (2007) were positive for this SNP.[74]
- E1b1a1a1g1a2 is defined by Z1725. This marker has been observed by The 1000 Genomes Project Consortium in Yoruba Nigerians and Luhya Kenyans.[86]
- E1b1a1a1g1c (YCC E1b1a4) is defined by M154. A Bamilike population tested 31.3% (15/48) for the marker. Bakaka speakers from Cameroon tested 8%.[45] An Ovimbundu test population found this SNP at 14% (14/100).[89] Members of this subclade have also been found in South Africa.[90][85]
- E1b1a1a1g1d is defined by V39. Trombetta et al. first published this SNP in 2011 but gave little population data about it.[5] It is only known to have been found in an African population.
E1b1a1a1h
E1b1a1a1h is defined by markers P268 and P269. It was first reported in a person from the Gambia.[91]
Phylogenetics
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E-P29 | 21 | III | 3A | 13 | Eu3 | H2 | B | E* | E | E | E | E | E | E | E | E | E | E |
| E-M33 | 21 | III | 3A | 13 | Eu3 | H2 | B | E1* | E1 | E1a | E1a | E1 | E1 | E1a | E1a | E1a | E1a | E1a |
| E-M44 | 21 | III | 3A | 13 | Eu3 | H2 | B | E1a | E1a | E1a1 | E1a1 | E1a | E1a | E1a1 | E1a1 | E1a1 | E1a1 | E1a1 |
| E-M75 | 21 | III | 3A | 13 | Eu3 | H2 | B | E2a | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 |
| E-M54 | 21 | III | 3A | 13 | Eu3 | H2 | B | E2b | E2b | E2b | E2b1 | - | - | - | - | - | - | - |
| E-P2 | 25 | III | 4 | 14 | Eu3 | H2 | B | E3* | E3 | E1b | E1b1 | E3 | E3 | E1b1 | E1b1 | E1b1 | E1b1 | E1b1 |
| E-M2 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a* | E3a | E1b1 | E1b1a | E3a | E3a | E1b1a | E1b1a | E1b1a | E1b1a1 | E1b1a1 |
| E-M58 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a1 | E3a1 | E1b1a1 | E1b1a1 | E3a1 | E3a1 | E1b1a1 | E1b1a1 | E1b1a1 | E1b1a1a1a | E1b1a1a1a |
| E-M116.2 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a2 | E3a2 | E1b1a2 | E1b1a2 | E3a2 | E3a2 | E1b1a2 | E1b1a2 | E1ba12 | removed | removed |
| E-M149 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a3 | E3a3 | E1b1a3 | E1b1a3 | E3a3 | E3a3 | E1b1a3 | E1b1a3 | E1b1a3 | E1b1a1a1c | E1b1a1a1c |
| E-M154 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a4 | E3a4 | E1b1a4 | E1b1a4 | E3a4 | E3a4 | E1b1a4 | E1b1a4 | E1b1a4 | E1b1a1a1g1c | E1b1a1a1g1c |
| E-M155 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a5 | E3a5 | E1b1a5 | E1b1a5 | E3a5 | E3a5 | E1b1a5 | E1b1a5 | E1b1a5 | E1b1a1a1d | E1b1a1a1d |
| E-M10 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a6 | E3a6 | E1b1a6 | E1b1a6 | E3a6 | E3a6 | E1b1a6 | E1b1a6 | E1b1a6 | E1b1a1a1e | E1b1a1a1e |
| E-M35 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b* | E3b | E1b1b1 | E1b1b1 | E3b1 | E3b1 | E1b1b1 | E1b1b1 | E1b1b1 | removed | removed |
| E-M78 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b1* | E3b1 | E1b1b1a | E1b1b1a1 | E3b1a | E3b1a | E1b1b1a | E1b1b1a | E1b1b1a | E1b1b1a1 | E1b1b1a1 |
| E-M148 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b1a | E3b1a | E1b1b1a3a | E1b1b1a1c1 | E3b1a3a | E3b1a3a | E1b1b1a3a | E1b1b1a3a | E1b1b1a3a | E1b1b1a1c1 | E1b1b1a1c1 |
| E-M81 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2* | E3b2 | E1b1b1b | E1b1b1b1 | E3b1b | E3b1b | E1b1b1b | E1b1b1b | E1b1b1b | E1b1b1b1 | E1b1b1b1a |
| E-M107 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2a | E3b2a | E1b1b1b1 | E1b1b1b1a | E3b1b1 | E3b1b1 | E1b1b1b1 | E1b1b1b1 | E1b1b1b1 | E1b1b1b1a | E1b1b1b1a1 |
| E-M165 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2b | E3b2b | E1b1b1b2 | E1b1b1b1b1 | E3b1b2 | E3b1b2 | E1b1b1b2a | E1b1b1b2a | E1b1b1b2a | E1b1b1b2a | E1b1b1b1a2a |
| E-M123 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b3* | E3b3 | E1b1b1c | E1b1b1c | E3b1c | E3b1c | E1b1b1c | E1b1b1c | E1b1b1c | E1b1b1c | E1b1b1b2a |
| E-M34 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b3a* | E3b3a | E1b1b1c1 | E1b1b1c1 | E3b1c1 | E3b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1b2a1 |
| E-M136 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3ba1 | E3b3a1 | E1b1b1c1a | E1b1b1c1a1 | E3b1c1a | E3b1c1a | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1b2a1a1 |
Research publications
The following research teams per their publications were represented in the creation of the YCC tree.
Phylogenetic trees
This phylogenetic tree of haplogroup subclades is based on the Y-Chromosome Consortium (YCC) 2008 Tree,[91] the ISOGG Y-DNA Haplogroup E Tree,[7] and subsequent published research.
- E1b1a1 (DYS271/M2/SY81, M291, P1/PN1, P189, P293, V43, V95, Z1101, Z1107, Z1116, Z1120, Z1122, Z1123, Z1124, Z1125, Z1127, Z1130, Z1133)[lower-alpha 6]
- E1b1a1a (L576)
- E1b1a1a1 (L86.1, L88.3, M180/P88, PAGES00066, P182, Z1111, Z1112)
- E1b1a1a1a (M58, PAGES00027)
- E1b1a1a1b (M116.2)
- E1b1a1a1c (M149)
- E1b1a1a1d (M155)
- E1b1a1a1e (M10, M66, M156, M195)
- E1b1a1a1f (L485)
- E1b1a1a1f1 (L514)
- E1b1a1a1f1a (M191/P86, P253/U247, U186, Z1712)
- E1b1a1a1f1a1 (P252/U174)
- E1b1a1a1f1a1a (P9.2)
- E1b1a1a1f1a1b (P115)
- E1b1a1a1f1a1c (P116)
- E1b1a1a1f1a1c1 (P113)
- E1b1a1a1f1a1d (Z1704)
- (L372)
- E1b1a1a1f1a1 (P252/U174)
- E1b1a1a1f1b (L515, L516, L517, M263.2)
- E1b1a1a1f1b1 (Z1893)
- (Z1894)
- E1b1a1a1f1b1 (Z1893)
- E1b1a1a1f1a (M191/P86, P253/U247, U186, Z1712)
- E1b1a1a1f1 (L514)
- E1b1a1a1g (U175)
- E1b1a1a1g1 (L220.3, L652, P277, P278.1, U209, M4254, M4230, CTS4921/M4243/V3224)
- E1b1a1a1g1a (U290)
- E1b1a1a1g1a1 (U181)
- E1b1a1a1g1a1a (L97)
- E1b1a1a1g1a2 (Z1725)
- E1b1a1a1g1a1 (U181)
- E1b1a1a1g1b (P59)
- E1b1a1a1g1c (M154)
- E1b1a1a1g1d (V39)
- E1b1a1a1g1a (U290)
- E1b1a1a1g1 (L220.3, L652, P277, P278.1, U209, M4254, M4230, CTS4921/M4243/V3224)
- E1b1a1a1h (P268, P269)
- E1b1a1a1 (L86.1, L88.3, M180/P88, PAGES00066, P182, Z1111, Z1112)
- E1b1a1a (L576)
- E1b1a1 (DYS271/M2/SY81, M291, P1/PN1, P189, P293, V43, V95, Z1101, Z1107, Z1116, Z1120, Z1122, Z1123, Z1124, Z1125, Z1127, Z1130, Z1133)[lower-alpha 6]
See also
Genetics
Y-DNA E subclades
Y-DNA backbone tree
Notes
- ↑ All were positive for U175.
- ↑ The publication refers to E-V38 as H22.
- ↑ E-M2 is approximately 7.7–7.9% of total US male population.
- ↑ The publication transposes M116.2 with M116.1 in Table 1.
- ↑ The YCAII STR marker value of 19–19 is also usually indicative of U175.
- ↑ DYS271/M2/SY81, P1/PN1, P189, P293, and M291 appear to form E1b1a1*. L576 forms a subclade immediately after the previously mentioned SNPs. L576 gave rise to a deeper subclade of M180/P88, P182, L88.3, L86, and PAGES0006. From this subclade, all the major subclades (i.e. E-U175 and E-L485) of E1b1a evolved. The exact position of V43 and V95 within these three subclades and E1b1a1a1b (M116.2), E1b1a1a1c (M149), and E1b1a1a1d (M155) remains uncertain.
References
- ↑ D'Atanasio E, Trombetta B, Bonito M, Finocchio A, Di Vito G, Seghizzi M (2018). "The peopling of the last Green Sahara revealed by high-coverage resequencing of trans-Saharan patrilineages.". Genome Biol 19 (1): 20. doi:10.1186/s13059-018-1393-5. PMID 29433568.
- ↑ 2.0 2.1 "E-M2 YTree". https://www.yfull.com/tree/E-M2/.
- ↑ 3.0 3.1 Trombetta, Beniamino (2015). "Phylogeographic Refinement and Large Scale Genotyping of Human Y Chromosome Haplogroup E Provide New Insights into the Dispersal of Early Pastoralists in the African Continent". Genome Biology and Evolution (Genome Biol Evol) 7 (7): 1940–1950. doi:10.1093/gbe/evv118. PMID 26108492.
- ↑ 4.0 4.1 4.2 Shriner, Daniel; Rotimi, Charles (2018). "Whole-Genome-Sequence-Based Haplotypes Reveal Single Origin of the Sickle Allele during the Holocene Wet Phase". American Journal of Human Genetics (Am J Hum Genet) 102 (4): 547–556. doi:10.1016/j.ajhg.2018.02.003. PMID 29526279.
- ↑ 5.0 5.1 5.2 MacAulay, Vincent, ed (January 2011). "A new topology of the human Y chromosome haplogroup E1b1 (E-P2) revealed through the use of newly characterized binary polymorphisms". PLOS ONE 6 (1): e16073. doi:10.1371/journal.pone.0016073. PMID 21253605. Bibcode: 2011PLoSO...616073T.
- ↑ "E-V43 YTree". https://www.yfull.com/tree/E-V43/.
- ↑ 7.0 7.1 International Society of Genetic Genealogy (3 February 2010). "Y-DNA Haplogroup E and its Subclades – 2010". http://www.isogg.org/tree/ISOGG_HapgrpE.html.
- ↑ Adams, Jonathan. "Africa During the Last 150,000 Years". http://www.esd.ornl.gov/projects/qen/nercAFRICA.html.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 9.7 "The Bantu expansion revisited: a new analysis of Y chromosome variation in Central Western Africa". Molecular Ecology 20 (13): 2693–708. July 2011. doi:10.1111/j.1365-294X.2011.05130.x. PMID 21627702.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 "Y-chromosomal diversity in the population of Guinea-Bissau: a multiethnic perspective". BMC Evolutionary Biology 7: 124. July 2007. doi:10.1186/1471-2148-7-124. PMID 17662131.
- ↑ 11.00 11.01 11.02 11.03 11.04 11.05 11.06 11.07 11.08 11.09 11.10 11.11 "Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes". European Journal of Human Genetics 13 (7): 867–76. July 2005. doi:10.1038/sj.ejhg.5201408. PMID 15856073.
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 "The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations". Annals of Human Genetics 65 (Pt 1): 43–62. January 2001. doi:10.1046/j.1469-1809.2001.6510043.x. PMID 11415522.
- ↑ 13.0 13.1 13.2 Wang, Ke (2020). "Ancient genomes reveal complex patterns of population movement, interaction, and replacement in sub-Saharan Africa". Science Advances 6 (24): eaaz0183. doi:10.1126/sciadv.aaz0183. ISSN 2375-2548. OCLC 8616876709. PMID 32582847. Bibcode: 2020SciA....6..183W.
- ↑ 14.0 14.1 14.2 Wang, Ke (2020). "Supplementary Materials for Ancient genomes reveal complex patterns of population movement, interaction, and replacement in sub-Saharan Africa". Science Advances 6 (24): eaaz0183. doi:10.1126/sciadv.aaz0183. ISSN 2375-2548. OCLC 8616876709. PMID 32582847. PMC 7292641. Bibcode: 2020SciA....6..183W. https://advances.sciencemag.org/content/suppl/2020/06/08/6.24.eaaz0183.DC1/aaz0183_SM.pdf.
- ↑ 15.0 15.1 15.2 Hawass, Zahi (2012). "Revisiting the harem conspiracy and death of Ramesses III: anthropological, forensic, radiological, and genetic study". British Medical Journal 345: e8268. doi:10.1136/bmj.e8268. ISSN 0959-8138. OCLC 825973553. PMID 23247979. https://www.researchgate.net/publication/233941674.
- ↑ Gad, Yehia Z (2021). "Insights from ancient DNA analysis of Egyptian human mummies: clues to disease and kinship". Human Molecular Genetics 30 (R1): R24–R28. doi:10.1093/hmg/ddaa223. ISSN 0964-6906. OCLC 8681412353. PMID 33059357.
- ↑ Prendergast, Mary E. (30 May 2019). "Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa". Science 365 (6448). doi:10.1126/science.aaw6275. ISSN 0036-8075. OCLC 8176642048. PMID 31147405. Bibcode: 2019Sci...365.6275P.
- ↑ Prendergast, Mary E. (30 May 2019). "Supplementary Materials for Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa". Science 365 (6448). doi:10.1126/science.aaw6275. ISSN 0036-8075. OCLC 8176642048. PMID 31147405. PMC 6827346. Bibcode: 2019Sci...365.6275P. https://science.sciencemag.org/content/sci/suppl/2019/05/29/science.aaw6275.DC1/aaw6275_Prendergast_SM.pdf.
- ↑ 19.0 19.1 19.2 19.3 19.4 19.5 19.6 19.7 Brielle, Esther S. (March 29, 2023). "Supplementary Data Files for Entwined African and Asian genetic roots of medieval peoples of the Swahili coast". Nature 615 (7954): 866–873. doi:10.1038/s41586-023-05754-w. ISSN 0028-0836. OCLC 9819552636. PMID 36991187. Bibcode: 2023Natur.615..866B.
- ↑ Barquera, Rodrigo (8 June 2020). "Origin and Health Status of First-Generation Africans from Early Colonial Mexico". Current Biology 30 (11): 2078–2091. doi:10.1016/j.cub.2020.04.002. ISSN 0960-9822. OCLC 8586564917. PMID 32359431.
- ↑ Fortes-Lima, Cesar A. (22 Nov 2021). "Disentangling the Impact of the Transatlantic Slave Trade in African Diaspora Populations from a Genomic Perspective". Africa, the Cradle of Human Diversity Cultural and Biological Approaches to Uncover African Diversity. Brill. pp. 305, 308–321. doi:10.1163/9789004500228_012. ISBN 978-90-04-50022-8. OCLC 1284920909. https://brill.com/view/book/9789004500228/BP000022.xml?body=pdf-43180.
- ↑ Peyroteo-Stjerna, Rita (21 February 2022). "Multidisciplinary investigation reveals an individual of West African origin buried in a Portuguese Mesolithic shell midden four centuries ago". Journal of Archaeological Science: Reports 42: 103370. doi:10.1016/j.jasrep.2022.103370. OCLC 1337974923.
- ↑ 23.0 23.1 23.2 23.3 23.4 Sandoval-Velasco, Marcela (7 September 2023). "The ancestry and geographical origins of St Helena's liberated Africans". American Journal of Human Genetics 110 (9): 1590–1599. doi:10.1016/j.ajhg.2023.08.001. ISSN 0002-9297. OCLC 9998699240. PMID 37683613. PMC 10502851. https://www.cell.com/ajhg/pdf/S0002-9297(23)00277-X.pdf.
- ↑ Isable, Kendra Briana (August 2021). "Contributing to the Discussion: The Health of Enslaved Africans Through the Lens of Bioarchaeology". California State University, Northridge. p. 34. https://scholarworks.calstate.edu/downloads/hd76s522x.
- ↑ Fortes-Lima, Cesar; Verdu, Paul (17 December 2020). "Anthropological genetics perspectives on the transatlantic slave trade". Human Molecular Genetics 30 (R1): R79–R87. doi:10.1093/hmg/ddaa271. PMID 33331897. https://academic.oup.com/hmg/article/30/R1/R79/6040749.
- ↑ Callaway, Ewen (December 7, 2016). "What DNA reveals about St Helena's freed slaves". Nature 540 (7632): 184–187. doi:10.1038/540184a.
- ↑ 27.00 27.01 27.02 27.03 27.04 27.05 27.06 27.07 27.08 27.09 27.10 27.11 27.12 27.13 27.14 27.15 27.16 27.17 27.18 27.19 Sandoval-Velasco, Marcela (7 September 2023). "Data S1. Tables S1–S13: The ancestry and geographical origins of St Helena's liberated Africans". American Journal of Human Genetics 110 (9): 1590–1599. doi:10.1016/j.ajhg.2023.08.001. ISSN 0002-9297. OCLC 9998699240. PMID 37683613.
- ↑ Fleskes, Raquel E. (2019). "Ancient DNA and bioarchaeological perspectives on European and African diversity and relationships on the colonial Delaware frontier". American Journal of Physical Anthropology 170 (2): 232–245. doi:10.1002/ajpa.23887. PMID 31270812. https://par.nsf.gov/servlets/purl/10110204.
- ↑ Racimo, Fernando (2020). "Beyond broad strokes: sociocultural insights from the study of ancient genomes". Nature Reviews Genetics 21 (6): 355–366. doi:10.1038/s41576-020-0218-z. ISSN 1471-0056. PMID 32127690. https://www.repository.cam.ac.uk/bitstream/handle/1810/302954/Review%20ancient%20DNA%20-%20Fernando%2c%20Martin%2c%20Marc%2c%20Hannes%2c%20Carles.pdf?sequence=1&isAllowed=y.
- ↑ Callaway, Ewen (November 5, 2019). "Genomes trace origins of enslaved people who died on remote island". Nature 575 (18): 18. doi:10.1038/d41586-019-03152-9. PMID 31690869. Bibcode: 2019Natur.575...18C. https://www.nature.com/articles/d41586-019-03152-9.
- ↑ Abel, Sarah; Schroeder, Hannes (October 2020). "From Country Marks to DNA Markers: The Genomic Turn in the Reconstruction of African Identities". Current Anthropology 61: S206. doi:10.1086/709550. https://www.journals.uchicago.edu/doi/epdf/10.1086/709550.
- ↑ 32.0 32.1 32.2 Olalde, Iñigo (2019). "Supplementary Materials for The genomic history of the Iberian Peninsula over the past 8000 years". Science 363 (6432): 1230–1234. doi:10.1126/science.aav4040. ISSN 0036-8075. OCLC 8024095449. PMID 30872528. PMC 6436108. Bibcode: 2019Sci...363.1230O. https://science.sciencemag.org/content/sci/suppl/2019/03/13/363.6432.1230.DC1/aav4040_Olalde_SM.pdf.
- ↑ 33.0 33.1 Olalde, Iñigo (2019). "The genomic history of the Iberian Peninsula over the past 8000 years, TablesS1-S5". Science 363 (6432): 1230–1234. doi:10.1126/science.aav4040. ISSN 0036-8075. OCLC 8024095449. PMID 30872528. Bibcode: 2019Sci...363.1230O.
- ↑ 34.0 34.1 Olalde, Iñigo (2019). "Materials/Methods, Supplementary Text, Tables, Figures, and/or References". Science 363 (6432): 1230–1234. doi:10.1126/science.aav4040. ISSN 0036-8075. OCLC 8024095449. PMID 30872528. Bibcode: 2019Sci...363.1230O.
- ↑ Fleskes, Raquel E. (June 5, 2023). "Historical genomes elucidate European settlement and the African diaspora in Delaware". Current Biology 33 (11): 2350–2358.e7. doi:10.1016/j.cub.2023.04.069. ISSN 0960-9822. OCLC 9874997102. PMID 37207647. https://www.researchgate.net/publication/370864951.
- ↑ Harney, Éadaoin (2023). "The genetic legacy of African Americans from Catoctin Furnace". Science 381 (500): eade4995. doi:10.1126/science.ade4995. PMID 37535739. https://reich.hms.harvard.edu/sites/reich.hms.harvard.edu/files/inline-files/Catoctin_main_Science.pdf.
- ↑ Harney, Éadaoin (2023). "Supplementary Materials for The genetic legacy of African Americans from Catoctin Furnace". Science 381 (500): eade4995. doi:10.1126/science.ade4995. PMID 37535739. https://reich.hms.harvard.edu/sites/reich.hms.harvard.edu/files/inline-files/Harney-etal-Catoctin-Science-AcceptedVersion_SM.pdf.
- ↑ 38.00 38.01 38.02 38.03 38.04 38.05 38.06 38.07 38.08 38.09 38.10 Harney, Éadaoin (2023). "Tables S1 to S24, S1.1, S3.2 to S3.7, and S4.1 for The genetic legacy of African Americans from Catoctin Furnace". Science 381 (500): eade4995. doi:10.1126/science.ade4995. PMID 37535739. https://www.science.org/doi/suppl/10.1126/science.ade4995/suppl_file/science.ade4995_tables_s1_to_s24_s1.1_s3.2_to_s3.7_and_s4.1.zip.
- ↑ 39.00 39.01 39.02 39.03 39.04 39.05 39.06 39.07 39.08 39.09 39.10 39.11 39.12 39.13 39.14 39.15 39.16 39.17 39.18 Fleskes, Raquel E. (2023). "Community-engaged ancient DNA project reveals diverse origins of 18th-century African descendants in Charleston, South Carolina". Anthropology 120 (3): e2201620120. doi:10.1073/pnas.2201620120. PMID 36623185. PMC 9934026. https://www.researchgate.net/publication/366977143.
- ↑ 40.0 40.1 40.2 40.3 40.4 40.5 Shriner, Daniel; Rotimi, Charles N. (2018). "Whole-Genome-Sequence-Based Haplotypes Reveal Single Origin of the Sickle Allele during the Holocene Wet Phase". American Journal of Human Genetics 102 (4): 547–556. doi:10.1016/j.ajhg.2018.02.003. ISSN 0002-9297. OCLC 7352712531. PMID 29526279.
- ↑ 41.0 41.1 41.2 Esoh, Kevin; Wonkam, Ambroise (2021). "Evolutionary history of sickle-cell mutation: implications for global genetic medicine". Human Molecular Genetics 30 (R1): R119–R128. doi:10.1093/hmg/ddab004. ISSN 0964-6906. OCLC 8885008275. PMID 33461216.
- ↑ Laval, Guillaume (2019). "Recent Adaptive Acquisition by African Rainforest Hunter-Gatherers of the Late Pleistocene Sickle-Cell Mutation Suggests Past Differences in Malaria Exposure". The American Journal of Human Genetics 104 (3): 553–561. doi:10.1016/j.ajhg.2019.02.007. ISSN 0002-9297. OCLC 8015758034. PMID 30827499.
- ↑ 43.0 43.1 43.2 43.3 Yaseen, Noor Taha (2020). "Sickle ß-globin haplotypes among patients with sickle cell anemia in Basra, Iraq: A cross-sectional study". Iraqi Journal of Hematology 9 (1): 23–29. doi:10.4103/ijh.ijh_20_19. ISSN 2072-8069. OCLC 8663256900. http://www.ijhonline.org/article.asp?issn=2072-8069;year=2020;volume=9;issue=1;spage=23;epage=29;aulast=Yaseen.
- ↑ 44.0 44.1 44.2 44.3 44.4 44.5 44.6 44.7 44.8 "The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations". American Journal of Human Genetics 74 (3): 532–44. March 2004. doi:10.1086/382286. PMID 14973781.
- ↑ 45.0 45.1 45.2 45.3 45.4 "A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes". American Journal of Human Genetics 70 (5): 1197–214. May 2002. doi:10.1086/340257. PMID 11910562.
- ↑ International HapMap Consortium (October 2005). "A haplotype map of the human genome". Nature 437 (7063): 1299–320. doi:10.1038/nature04226. PMID 16255080. Bibcode: 2005Natur.437.1299T.
- ↑ 47.0 47.1 47.2 47.3 "Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny". American Journal of Human Genetics 70 (1): 265–8. January 2002. doi:10.1086/338306. PMID 11719903.
- ↑ 48.0 48.1 48.2 "Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel". European Journal of Human Genetics 18 (8): 915–23. August 2010. doi:10.1038/ejhg.2010.21. PMID 20234393.
- ↑ 49.0 49.1 "Genetic diversity on the Comoros Islands shows early seafaring as major determinant of human biocultural evolution in the Western Indian Ocean". European Journal of Human Genetics 19 (1): 89–94. January 2011. doi:10.1038/ejhg.2010.128. PMID 20700146.
- ↑ "Y-chromosome lineages in Cabo Verde Islands witness the diverse geographic origin of its first male settlers". Human Genetics 113 (6): 467–72. November 2003. doi:10.1007/s00439-003-1007-4. PMID 12942365.
- ↑ 51.0 51.1 51.2 51.3 "On the origins and admixture of Malagasy: new evidence from high-resolution analyses of paternal and maternal lineages". Molecular Biology and Evolution 26 (9): 2109–24. September 2009. doi:10.1093/molbev/msp120. PMID 19535740.
- ↑ 52.0 52.1 "High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males". European Journal of Human Genetics 13 (7): 856–66. July 2005. doi:10.1038/sj.ejhg.5201390. PMID 15756297.
- ↑ "Forensic data and microvariant sequence characterization of 27 Y-STR loci analyzed in four Eastern African countries". Forensic Science International. Genetics 27: 123–131. March 2017. doi:10.1016/j.fsigen.2016.12.015. PMID 28068531.
- ↑ Plaster et al. Y-DNA E subclades
- ↑ Plaster CA (2011-09-28). Variation in Y chromosome, mitochondrial DNA and labels of identity on Ethiopia. discovery.ucl.ac.uk (Doctoral). Retrieved 2018-06-27.
- ↑ "Deep into the roots of the Libyan Tuareg: a genetic survey of their paternal heritage". American Journal of Physical Anthropology 145 (1): 118–24. May 2011. doi:10.1002/ajpa.21473. PMID 21312181.
- ↑ "Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample". International Journal of Legal Medicine 122 (3): 251–5. May 2008. doi:10.1007/s00414-007-0203-5. PMID 17909833.
- ↑ 58.0 58.1 58.2 "High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula". American Journal of Human Genetics 68 (4): 1019–29. April 2001. doi:10.1086/319521. PMID 11254456.
- ↑ 59.0 59.1 "Phylogeographic analysis of haplogroup E3b (E-M215) y chromosomes reveals multiple migratory events within and out of Africa". American Journal of Human Genetics 74 (5): 1014–22. May 2004. doi:10.1086/386294. PMID 15042509.
- ↑ "Ancestral Asian source(s) of new world Y-chromosome founder haplotypes". American Journal of Human Genetics 64 (3): 817–31. March 1999. doi:10.1086/302282. PMID 10053017.
- ↑ 61.0 61.1 "A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa". American Journal of Human Genetics 75 (2): 338–45. August 2004. doi:10.1086/423147. PMID 15202071.
- ↑ "Y-chromosome variation among Sudanese: restricted gene flow, concordance with language, geography, and history". American Journal of Physical Anthropology 137 (3): 316–23. November 2008. doi:10.1002/ajpa.20876. PMID 18618658.
- ↑ . Branka Grskovic, Andro Vrdoljak, Maja Popovic, Ivica Valpotic, Simun Andelinovic, Vlastimil Stenzl, Edvard Ehler, Ludvik Urban, Gordana Lackovic, Peter Underhill, Dragan Primorac"Croatian national reference Y-STR haplotype database". Molecular Biology Reports 39 (7): 7727–41. July 2012. doi:10.1007/s11033-012-1610-3. PMID 22391654.
- ↑ "Population structure in the Mediterranean basin: a Y chromosome perspective". Annals of Human Genetics 70 (Pt 2): 207–25. March 2006. doi:10.1111/j.1529-8817.2005.00224.x. PMID 16626331.
- ↑ "Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography". European Journal of Human Genetics 12 (10): 855–63. October 2004. doi:10.1038/sj.ejhg.5201225. PMID 15280900.
- ↑ 66.0 66.1 "The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula". American Journal of Human Genetics 83 (6): 725–36. December 2008. doi:10.1016/j.ajhg.2008.11.007. PMID 19061982.
- ↑ Noora R. Al-Snan1; Safia A. Messaoudi; Yahya M. Khubrani; Jon H. Wetton; Mark A. Jobling; Moiz Bakhiet (2020). "Geographical structuring and low diversity of paternal lineages in Bahrain shown by analysis of 27 Y-STRs". Molecular Genetics and Genomics 295 (6): 1315–1324. doi:10.1007/s00438-020-01696-4. PMID 32588126.
- ↑ "Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions". BMC Genetics 10: 59. September 2009. doi:10.1186/1471-2156-10-59. PMID 19772609.
- ↑ 69.0 69.1 "Y-chromosome diversity characterizes the Gulf of Oman". European Journal of Human Genetics 16 (3): 374–86. March 2008. doi:10.1038/sj.ejhg.5201934. PMID 17928816.
- ↑ "Iran: tricontinental nexus for Y-chromosome driven migration". Human Heredity 61 (3): 132–43. 2006. doi:10.1159/000093774. PMID 16770078.
- ↑ "Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations". Molecular Phylogenetics and Evolution 28 (3): 458–72. September 2003. doi:10.1016/S1055-7903(03)00039-3. PMID 12927131.
- ↑ "Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan". European Journal of Human Genetics 15 (1): 121–6. January 2007. doi:10.1038/sj.ejhg.5201726. PMID 17047675.
- ↑ "Excavating Y-chromosome haplotype strata in Anatolia". Human Genetics 114 (2): 127–48. January 2004. doi:10.1007/s00439-003-1031-4. PMID 14586639.
- ↑ 74.0 74.1 74.2 74.3 "Sub-populations within the major European and African derived haplogroups R1b3 and E3a are differentiated by previously phylogenetically undefined Y-SNPs". Human Mutation 28 (1): 97. January 2007. doi:10.1002/humu.9469. PMID 17154278.
- ↑ "Genetic origin, admixture, and asymmetry in maternal and paternal human lineages in Cuba". BMC Evolutionary Biology 8: 213. July 2008. doi:10.1186/1471-2148-8-213. PMID 18644108.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedBryc2010 - ↑ "Reconstructing the population history of Nicaragua by means of mtDNA, Y-chromosome STRs, and autosomal STR markers". American Journal of Physical Anthropology 143 (4): 591–600. December 2010. doi:10.1002/ajpa.21355. PMID 20721944.
- ↑ "Genetic make up and structure of Colombian populations by means of uniparental and biparental DNA markers". American Journal of Physical Anthropology 143 (1): 13–20. September 2010. doi:10.1002/ajpa.21270. PMID 20734436.
- ↑ "Analysis of Y chromosome SNPs in Alagoas, Northeastern Brazil". Forensic Science International: Genetics Supplement Series 2 (1): 421–422. December 2009. doi:10.1016/j.fsigss.2009.08.166.
- ↑ "The Africa male lineages of Bahia's people—Northeast Brazil: A preliminary SNPs study". Forensic Science International: Genetics Supplement Series 2 (1): 349–350. December 2009. doi:10.1016/j.fsigss.2009.07.010.
- ↑ Tanya M Simms 2011, The Peopling of the Bahamas: A Phylogeographical Perspective pg. 194
- ↑ 82.0 82.1 "Niger-Congo speaking populations and the formation of the Brazilian gene pool: mtDNA and Y-chromosome data". American Journal of Physical Anthropology 133 (2): 854–67. June 2007. doi:10.1002/ajpa.20604. PMID 17427922.
- ↑ 83.0 83.1 "Y-chromosomal variation in sub-Saharan Africa: insights into the history of Niger-Congo groups". Molecular Biology and Evolution 28 (3): 1255–69. March 2011. doi:10.1093/molbev/msq312. PMID 21109585.
- ↑ 84.0 84.1 84.2 84.3 "Little genetic differentiation as assessed by uniparental markers in the presence of substantial language variation in peoples of the Cross River region of Nigeria". BMC Evolutionary Biology 10: 92. March 2010. doi:10.1186/1471-2148-10-92. PMID 20356404.
- ↑ 85.0 85.1 "Development of a single base extension method to resolve Y chromosome haplogroups in sub-Saharan African populations". Investigative Genetics 1 (1): 6. September 2010. doi:10.1186/2041-2223-1-6. PMID 21092339.
- ↑ 86.0 86.1 "A map of human genome variation from population-scale sequencing". Nature 467 (7319): 1061–73. October 2010. doi:10.1038/nature09534. PMID 20981092. Bibcode: 2010Natur.467.1061T.
- ↑ Reynolds, David; Squecco, Adriano. "Y-Chromosome Genome Comparison". http://www.daver.info/ysub/.
- ↑ "The imprint of the Slave Trade in an African American population: mitochondrial DNA, Y chromosome and HTLV-1 analysis in the Noir Marron of French Guiana". BMC Evolutionary Biology 10: 314. October 2010. doi:10.1186/1471-2148-10-314. PMID 20958967.
- ↑ "Y-SNP analysis in an Angola population". Forensic Science International: Genetics Supplement Series 3 (1): e369–e370. December 2011. doi:10.1016/j.fsigss.2011.09.046.
- ↑ "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–61. November 2000. doi:10.1038/81685. PMID 11062480.
- ↑ 91.0 91.1 "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–8. May 2008. doi:10.1101/gr.7172008. PMID 18385274.
Sources for conversion tables
- Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H. et al. (February 2001). "A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania". The American Journal of Human Genetics 68 (2): 432–443. doi:10.1086/318205. PMID 11170891.
- Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi et al. (1 July 2001). "Hierarchical Patterns of Global Human Y-Chromosome Diversity". Molecular Biology and Evolution 18 (7): 1189–1203. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- Jobling, Mark A.; Tyler-Smith, Chris (2000), "New uses for new haplotypes", Trends in Genetics 16 (8): 356–62, doi:10.1016/S0168-9525(00)02057-6, PMID 10904265
- Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora et al. (February 2001). "Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages". European Journal of Human Genetics 9 (2): 97–104. doi:10.1038/sj.ejhg.5200597. PMID 11313742.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William et al. (September 2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics 69 (3): 615–628. doi:10.1086/323299. PMID 11481588.
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science 290 (5494): 1155–9, doi:10.1126/science.290.5494.1155, PMID 11073453, Bibcode: 2000Sci...290.1155S
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics 65 (6): 1718–1724. doi:10.1086/302680. PMID 10577926.
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–361. doi:10.1038/81685. PMID 11062480.
External links
- Haplogroup E1b1a FTDNA Project
- Distribution of E1b1a/E3a in Africa
- Spread of Haplogroup E3a, from National Geographic
 |


