Biology:Haplogroup H (mtDNA)
| Haplogroup H | |
|---|---|
| Possible time of origin | 20,000–25,000 YBP |
| Possible place of origin | Southwest Asia (West Asia or Western Asia), Lesser Caucasus |
| Ancestor | HV[1] |
| Descendants | H* lineages; subclades H1, H2, H3, H4, H5'36, H6, H7, H8, H9, H10, H11, H12, H13, H14, H15, H16, H18, H19, H20, H22, H23, H24, H25, H26, H28, H29, H31, H32, H33, H34, H35, H37, H38, H39, 16129(H17+H27), 16129(H21+H30) (numbers to H144)[2] |
| Defining mutations | G2706A, T7028C[3] |
Haplogroup H is a human mitochondrial DNA (mtDNA) haplogroup. The clade is believed to have originated in Southwest Asia, near present day Syria,[1] around 20,000 to 25,000 years ago. Mitochondrial haplogroup H is today predominantly found in Europe, and is believed to have evolved before the Last Glacial Maximum (LGM). It first expanded in the northern Near East and Southern Caucasus soon, and later migrations from Iberia suggest that the clade reached Europe before the Last Glacial Maximum. The haplogroup has also spread to parts of Africa, Siberia and Inner Asia. Today, around 40% of all maternal lineages in Europe belong to haplogroup H.
Origin
Haplogroup H is a descendant of haplogroup HV. The Cambridge Reference Sequence (CRS), which until recently was the human mitochondrial sequence to which all others were compared, belongs to haplogroup H2a2a1 (human mitochondrial sequences should now be compared with the ancestral Reconstructed Sapiens Reference Sequence (RSRS)).[4] Several independent studies conclude that haplogroup H probably evolved in Western Asia c. 25,000 years ago.
In July 2008 ancient mtDNA from an individual called Paglicci 23, whose remains were dated to 28,000 years ago and excavated from Paglicci Cave (Apulia, Italy), were found to be identical to the Cambridge Reference Sequence in HVR1.[5] This once was believed to indicate haplogroup H, but researchers now recognize that CRS HVR1 also appears in U or HV, because there are no HVR1 mutations that separate CRS from the haplogroup R founder. Haplogroup HV derives from the haplogroup R0 which in turn derives from haplogroup R is a descendant of macro-haplogroup N like its sibling M, is a descendant of haplogroup L3.
MtDNA H had frequency of 19% among Neolithic Early European Farmers and virtually absent among Mesolithic European hunter gatherers.[6]
MtDNA H was also present in the Cucuteni–Trypillia culture.[7]
The clade has been observed among ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from the pre-Ptolemaic/late New Kingdom and Ptolemaic periods.[8]
Additionally, haplogroup H has been found among specimens at the mainland cemetery in Kulubnarti, Sudan, which date from the Early Christian period (AD 550–800).[9]
Distribution
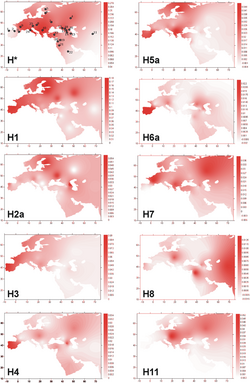
Haplogroup H is the most common mtDNA clade in Europe.[10] It is found in approximately 41% of native Europeans.[11][12] The lineage is also common in North Africa and the Middle East.[13]
The majority of the European populations have an overall haplogroup H frequency of 40–50%, with frequencies decreasing in the southeast. The clade reaches 20% in the Near East and Caucasus, 17% in Iran, and <10% in the Arabian Peninsula, Northern India and Central Asia.[1][14]
Undifferentiated haplogroup H has been found among Palestinians (14%),[15] Syrians (13.6%),[15] Druze (10.6%),[15] Iraqis (9.5%),[15] Somalis (6.7%),[15] Egyptians (5.7% in El-Hayez;[16] 14.7% in Gurna[17]), Saudis (5.3–10%),[15] Soqotri (3.1%),[18] Nubians (1.3%),[15] and Yemenis (0–13.9%).[15]
Subclades
Among all these clades, the subhaplogroups H1 and H3 have been subject to a more detailed study and would be associated to the Magdalenian expansion from SW Europe c. 13,000 years ago:[19]
H1
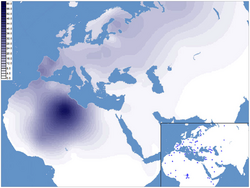
H1 encompasses an important fraction of Western European mtDNA lineages, reaching its local peak among contemporary Basques (27.8%). The clade also occurs at high frequencies elsewhere in the Iberian Peninsula, as well as in the Maghreb (Tamazgha). The haplogroup frequency is above 10% in many other parts of Europe (France, Sardinia, parts of the British Isles, Alps, large portions of Eastern Europe), and surpasses 5% in nearly all of the continent.[1] Its H1b subclade is most common in eastern Europe and NW Siberia.[20]
As of 2010[update], the highest frequency of the H1 subclade has been found among the Tuareg inhabiting the Fezzan region in Libya (61%).[21] The basal H1* haplogroup is found among the Tuareg inhabiting the Gossi area in Mali (4.76%).[22]
The rare H1cb subclade is concentrated among Fulani groups inhabiting the Sahel.[23]
Haplogroup H has been found in various fossils that were analysed for ancient DNA, including specimens associated with the Linearbandkeramik culture (H1e, Halberstadt-Sonntagsfeld, 1/22 or ~5%; H1 or H1au1b, Karsdorf, 1/2 or 50%), Germany Middle Neolithic (H1e1a, Esperstedt, 1/1 or 100%), Iberia Early Neolithic (H1, El Prado de Pancorbo, 1/2 or 50%), Iberia Middle Neolithic (H1, La Mina, 1/4 or 25%), and Iberia Chalcolithic (H1t, El Mirador Cave, 1/12 or ~8%).[24] Haplogroup H has been observed in ancient Guanche fossils excavated in Gran Canaria and Tenerife on the Canary Islands, which have been radiocarbon-dated to between the 7th and 11th centuries CE. At the Tenerife site, these clade-bearing individuals were found to belong to the H1cf subclade (1/7; ~14%); at the Gran Canaria site, the specimens carried the H2a subhaplogroup (1/4; 25%).[25] Additionally, ancient Guanche (Bimbaches) individuals excavated in Punta Azul, El Hierro, Canary Islands were all found to belong to the H1 maternal subclade. These locally born individuals were dated to the 10th century and carried the H1-16260 haplotype, which is exclusive to the Canary Islands and Algeria.[26]
- Frequencies of haplogroup H1 in the world (Ottoni et al. 2010)
| Region or Population | H1% | No. of subjects |
|---|---|---|
| Africa | ||
| Libyan Tuareg | 61 | 129 |
| Tuareg (West Sahel) | 23.3 | 90 |
| Berbers (Morocco) | 20.2 | 217 |
| Morocco | 12.2 | 180 |
| Berbers (Tunisia) | 13.4 | 276 |
| Tunisia | 10.6 | 269 |
| Mozabite | 9.8 | 80 |
| Siwas (Egypt) | 1.1 | 184 |
| Western Sahara | 14.8 | 128 |
| Mauritania | 6.9 | 102 |
| Senegal | 0 | 100 |
| Fulani (Chad–Cameroon) | 0 | 186 |
| Cameroon | 0 | 142 |
| Chad | 0 | 77 |
| Buduma (Niger) | 0 | 30 |
| Nigeria | 0 | 69 |
| Ethiopia | 0 | 82 |
| Amhara (Ethiopia) | 0 | 90 |
| Oromo (Ethiopia) | 0 | 117 |
| Sierra Leone | 0 | 155 |
| Guineans (Guiné Bissau) | 0 | 372 |
| Mali | 0 | 83 |
| Kikuyu (Kenya) | 0 | 24 |
| Benin | 0 | 192 |
| Asia | ||
| Central Asia | 0.7 | 445 |
| Pakistan | 0 | 100 |
| Yakuts | 1.7 | 58 |
| Caucasus | ||
| Caucasus (north) | 8.8 | 68 |
| Caucasus (south) | 2.3 | 132 |
| Northwestern Caucasus | 4.7 | 234 |
| Armenians | 2.3 | 175 |
| Daghestan | 2.5 | 269 |
| Georgians | 1 | 193 |
| Karachay-Balkars | 4.4 | 203 |
| Ossetians | 2.4 | 296 |
| Europe | ||
| Andalusia | 24.3 | 103 |
| Basques (Spain) | 27.8 | 108 |
| Catalonia | 13.9 | 101 |
| Galicia | 17.7 | 266 |
| Pasiegos (Cantabria) | 23.5 | 51 |
| Portugal | 25.5 | 499 |
| Spain (miscellaneous) | 18.9 | 132 |
| Italy (north) | 11.5 | 322 |
| Italy (center) | 6.3 | 208 |
| Italy (south) | 8.7 | 206 |
| Sardinia | 17.9 | 106 |
| Sicily | 10 | 90 |
| Finland | 18 | 78 |
| Volga-Ural Finnic speakers | 13.6 | 125 |
| Basques (France) | 17.5 | 40 |
| Béarnaise | 14.8 | 27 |
| France | 12.3 | 106 |
| Estonia | 16.7 | 114 |
| Saami | 0 | 57 |
| Lithuania | 1.7 | 180 |
| Hungary | 11.3 | 303 |
| Czech Republic | 10.8 | 102 |
| Ukraine | 9.9 | 191 |
| Poland | 9.3 | 86 |
| Russia | 13.5 | 312 |
| Austria | 10.6 | 2487 |
| Germany | 6 | 100 |
| Romania | 9.4 | 360 |
| Netherlands | 8.8 | 34 |
| Greece (Aegean islands) | 1.6 | 247 |
| Greece (mainland) | 6.3 | 79 |
| Macedonia | 7.1 | 252 |
| Albania | 2.9 | 105 |
| Turks | 3.3 | 360 |
| Balkans | 5.4 | 111 |
| Croatia | 8.3 | 84 |
| Slovaks | 7.6 | 119 |
| Slovak (East) | 16.8 | 137 |
| Slovak (West) | 14.2 | 70 |
| Middle East | ||
| Arabian Peninsula | 0 | 94 |
| Arabian Peninsula (incl. Yemen, Oman) | 0.8 | 493 |
| Druze | 3.4 | 58 |
| Dubai (United Arab Emirates) | 0.4 | 249 |
| Iraq | 1.9 | 206 |
| Jordanians | 1.7 | 173 |
| Lebanese | 4.2 | 167 |
| Syrians | 0 | 159 |
H3
H3 is found throughout the whole of Europe and in the Maghreb,[1] and is believed to have originated among Mesolithic hunter-gatherers in south-western Europe between 9 000 and 11 000 years ago. H3 represents the second largest fraction of the H genome after H1 and has a somewhat similar distribution, with peaks in Portugal, Spain, Scandinavia and Finland. It is common in Portugal (12%), Sardinia (11%), Galicia (10%), the Basque country (10%), Ireland (6%), Norway (6%), Hungary (6%) and southwestern France (5%).[1][27][28] Studies have suggested haplogroup H3 is highly protective against AIDS progression.[29]
Example of H3 sub-groups are:[28]
- H3a and H3g, found in north-west Europe;
- H3b and H3k, found in the British Isles and Catalonia;
- H3c, found in Western Europe, including among the Basques;
- H3h, found throughout northern Europe, including the remains of Cerdic (519 to 534), King of Wessex;[30]
- H3i found in Ireland and Scotland;
- H3j found in Italy;
- H3v found especially in Germanic countries and;
- H3z found in Atlantic Europe.
The basal H3* haplogroup is found among the Tuareg inhabiting the Gossi area in Mali (4.76%).[22]
H5
H5 may have evolved in West Asia, where it is most frequent and diverse in the Western Caucasus. However, its H5a subclade has a stronger representation in Europe, though at low levels.[31]
H2, H6 and H8
The H2, H6 and H8 haplogroups are somewhat common in Eastern Europe and the Caucasus.[19] They may be the most common H subclades among Central Asians and have also been found in West Asia.[20] H2a5 has been found in the Basque Country, [32] and in Norway , Ireland and Slovakia.[3] H6a1a1a is common among Ashkenazi Jews.[33]
H4
H4 is often found in the Iberian peninsula,[32] Britain and Ireland at levels between 1-5% of the population. It is associated with Neolithic migrations.
H4 and H13, along with H2 account for 42% of the hg H lineages in Egypt.[34]
H7
The H7 subhaplogroup is present in both Europe and West Asia. Its subclade H7c1 is present in Druze people and in Saudi Arabia.[35] H7c2 is present in such peoples as Ashkenazi Jews, Sardinians, and Dutch people.[36] H7e is present in Ashkenazi Jews, Germans, Sardinians, and others.[37] H7a1b is found today in Scotland, England , Denmark , Finland and Sardinia.[38]
H9
H9 is present in Yemenis.[39] The subclade H9a exists in Welsh people,[40] Calabrians[41] and Crimean Karaites.[42][43] H9a samples were recovered from two ancient people in Lebanon.[44]
H10
Haplogroup H10 is subclade which came into existence between 6,300 and 10,900 years ago. Its descendant branches are H10a, H10b, H10c, H10d, H10e, H10f, H10g, and H10h.[45]
Haplogroup H10e has been found at a neolithic site, namely the Bom Santo cave near Lisbon. This is the oldest sample of H10 which has ever been found and it has been dated to 3735 BCE (+- 45 years).[46]
H11
H11 is commonly found in Central Europe.[32]
H12
Italians are notable carriers of H12 and its two branches. H12a has been detected in such regions as Sicily[47] and Calabria.[48]
H13
The H13 subhaplogroup is present in both Europe and West Asia. H13 is also found in the Caucasus; H13c was found in a 9,700 year old sample in Mesolithic Georgia[49] and H13a2a and H13a2b are found in Armenians in Armenia.[50][51]
H14
The root level of H14 is found in northwestern Europeans, such as in Ireland.[52]
Its subclade H14a is encountered among such populations as Armenians from Turkey,[53] Sardinians from Italy,[54] and Persian Jews and Iraqi Jews.[55] The branch H14a2 is present among Romani people from Spain [56] and Croats[57] and is common in Iran.[52]
Its subclade H14b has a presence in many European and West Asian populations including Assyrian people,[58] Persians[59] and Armenians from Iran,[60] and people in Tuscany (central Italy),[61] Switzerland, Germany, Ireland, Spain, and Qatar.[52] The branch H14b1 is especially prevalent in France but also found in neighboring Monaco and in Scotland.[52] The branch H14b3 has been found in Armenians from Artsakh[62] and people from Armenia, Turkey, Saudi Arabia, Italy, and Scotland.[52] H14b4 is in Italy and Germany.[52]
H15
H15 includes the base level and the branches H15a and H15b. H15a1b is present in Greeks.[63] H15b is present in Armenians, Druze, Ashkenazi Jews, Danes, and other peoples of Europe and the Near East.[64]
H16
H16 is encountered in numerous European populations, such as Norway and England .[52] Its subclade H16a is found in Czechs[65] and in Germany , Scotland, and the Netherlands.[52] H16a1 is similarly found in Europe, including in Denmark .[66] H16b is another common branch and among other places is present in Sardinia.[67] H16c was found in archaeological human remains from Iron Age cemetery in Lejasbitēni, Latvia,[68] it is currently found in Sweden, Great Britain, Lithuania, Poland , Germany , Latvia and elsewhere.[52] H16d is found in Italy, France , Ireland, England , and other parts of Europe.[52] H16e is especially common in Sweden.[52]
In July 2008 ancient mtDNA from an individual called Paglicci 23, whose remains were dated to 28,000 years ago and excavated from Paglicci Cave (Apulia, Italy), were found to be identical to the Cambridge Reference Sequence in HVR1.[5]
H17
H17 is especially prevalent in Ireland but also found in England , Scotland, Wales, Sweden, and Germany .[52] H17a is similarly found across Europe, such as in Sardinia[69] and Lithuania.[52]
H18
H18 occurs on the Arabian Peninsula. [70]
H20 and H21
These haplogroups are both found in the Caucasus region.[31] H20 also appears at low levels in the Iberian Peninsula (less than 1%), Arabian Peninsula (1%) and Near East (2%).[70]
H22 through H95a
These subclades are found mostly in Europe, South-West Asia and Central Asia.
H24 is found in Romance-speaking countries' populations (France , Spain , and Romani people from Spain)[71] as well as in Germany , Ireland, Scotland, and England .[52]
H45 is found in Ireland and Spain .[52] Its subclade H45a is especially found in Finland [72] but also found in Sweden.[52] H45b exists among multiple kinds of western Europeans, including people in Ireland,[73] Scotland, England , and Germany .[52]
H47 has been found in Syria, Italy, and Armenians and infrequently in Ashkenazi Jews.[74] Its subclade H47a is exclusively European, being found in such countries as England , Ireland, Czechia, and Bulgaria.[52]
H53 is encountered in such countries as Spain (including among Basques)[75] and Poland [76] and as far east as Xinjiang in western China (among Uyghurs).[77]
H69 is a European branch found in Finns,[78] Irish people,[79] and inhabitants of Sweden, Germany , and Switzerland .[52]
H91a is associated with the Uyghur ethnic minority of western China.[80]
H96 and above
These were the most recently discovered and named major branches of H.
H105 is found in Italy and Hungary.[52]
H106 is found in Italy, France , Austria, England , and Germany .[52]
H107 is encountered in Russia .[81]
Tree

This phylogenetic tree of haplogroup H subclades is based on Build 17 (February 2016) of the Phylotree, an internationally accepted standard.[82] The full tree can be viewed at Phylotree.
| mtDNA HG "H" p-tree |
|---|
|
Genetic traits
Haplogroup H was found as a possible increased risk factor for ischemic cardiomyopathy development.[83]
Popular culture
In his popular book The Seven Daughters of Eve, Bryan Sykes named the originator of this mtDNA haplogroup Helena. Stephen Oppenheimer uses the very similar name Helina in his book The Origins of the British.
See also
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 "The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool". American Journal of Human Genetics 75 (5): 910–8. November 2004. doi:10.1086/425590. PMID 15382008.
- ↑ "H144 MTree". https://www.yfull.com/mtree/H144/.
- ↑ 3.0 3.1 "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation 30 (2): E386–94. February 2009. doi:10.1002/humu.20921. PMID 18853457.
- ↑ "A "Copernican" reassessment of the human mitochondrial DNA tree from its root". American Journal of Human Genetics 90 (4): 675–84. April 2012. doi:10.1016/j.ajhg.2012.03.002. PMID 22482806.
- ↑ 5.0 5.1 "A 28,000 years old Cro-Magnon mtDNA sequence differs from all potentially contaminating modern sequences". PLOS ONE 3 (7): e2700. July 2008. doi:10.1371/journal.pone.0002700. PMID 18628960. Bibcode: 2008PLoSO...3.2700C.
- ↑ "Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans". Nature Communications 4: 1764. 2013. doi:10.1038/ncomms2656. PMID 23612305. Bibcode: 2013NatCo...4.1764..
- ↑ "Mitochondrial DNA analysis of eneolithic trypillians from Ukraine reveals neolithic farming genetic roots". PLOS ONE 12 (2): e0172952. 2017. doi:10.1371/journal.pone.0172952. PMID 28235025. Bibcode: 2017PLoSO..1272952N.
- ↑ "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications 8: 15694. May 2017. doi:10.1038/ncomms15694. PMID 28556824. Bibcode: 2017NatCo...815694S.
- ↑ "Abstract Book of the IUAES Inter-Congress 2016 – A community divided? Revealing the community genome(s) of Medieval Kulubnarti using next- generation sequencing". Abstract Book of the Iuaes Inter-Congress 2016 (IUAES): 115. 2016. https://bib.irb.hr/prikazi-rad?&lang=EN&rad=824672.
- ↑ "Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson's disease in Italians". European Journal of Human Genetics 13 (6): 748–52. June 2005. doi:10.1038/sj.ejhg.5201425. PMID 15827561.
- ↑ Sykes, Bryan (2001). The Seven Daughters of Eve. London; New York: Bantam Press. ISBN 978-0393020182.
- ↑ "Maternal Ancestry". Oxford Ancestors. http://www.oxfordancestors.com/content/view/35/55/.
- ↑ "Haplogroup H". Atlas of the Human Journey – The Genographic Project. National Geographic. https://genographic.nationalgeographic.com/genographic/atlas.html?card=mm024.
- ↑ "Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans". BMC Genetics 5: 26. August 2004. doi:10.1186/1471-2156-5-26. PMID 15339343.
- ↑ 15.0 15.1 15.2 15.3 15.4 15.5 15.6 15.7 Non, Amy. "Analyses if Genetic Data Within A=an Interdisciplinary Framework to Investigate Recent Human Evolutionary History and Complex Disease". University of Florida. http://etd.fcla.edu/UF/UFE0041981/non_a.pdf.
- ↑ "Near eastern neolithic genetic input in a small oasis of the Egyptian Western Desert". American Journal of Physical Anthropology 140 (2): 336–46. October 2009. doi:10.1002/ajpa.21078. PMID 19425100.
- ↑ "Mitochondrial DNA sequence diversity in a sedentary population from Egypt". Annals of Human Genetics 68 (Pt 1): 23–39. January 2004. doi:10.1046/j.1529-8817.2003.00057.x. PMID 14748828.
- ↑ "Out of Arabia-the settlement of island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity". American Journal of Physical Anthropology 138 (4): 439–47. April 2009. doi:10.1002/ajpa.20960. PMID 19012329.
- ↑ 19.0 19.1 "High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium". Genome Research 15 (1): 19–24. January 2005. doi:10.1101/gr.3182305. PMID 15632086.
- ↑ 20.0 20.1 "Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia". Molecular Biology and Evolution 21 (11): 2012–21. November 2004. doi:10.1093/molbev/msh209. PMID 15254257.
- ↑ "Mitochondrial haplogroup H1 in north Africa: an early holocene arrival from Iberia". PLOS ONE 5 (10): e13378. October 2010. doi:10.1371/journal.pone.0013378. PMID 20975840. Bibcode: 2010PLoSO...513378O.
- ↑ 22.0 22.1 "Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel". European Journal of Human Genetics 18 (8): 915–23. August 2010. doi:10.1038/ejhg.2010.21. PMID 20234393.
- ↑ "Internal diversification of non-Sub-Saharan haplogroups in Sahelian populations and the spread of pastoralism beyond the Sahara". American Journal of Physical Anthropology 164 (2): 424–434. October 2017. doi:10.1002/ajpa.23285. PMID 28736914.
- ↑ "Parallel palaeogenomic transects reveal complex genetic history of early European farmers". Nature 551 (7680): 368–372. November 2017. doi:10.1038/nature24476. PMID 29144465. Bibcode: 2017Natur.551..368L.
- ↑ "Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modern North Africans". Current Biology 27 (21): 3396–3402.e5. November 2017. doi:10.1016/j.cub.2017.09.059. PMID 29107554.
- ↑ "Genetic studies on the prehispanic population buried in Punta Azul cave (El Hierro, Canary Islands)". Journal of Archaeological Science 78: 20–28. 2017. doi:10.1016/j.jas.2016.11.004. Bibcode: 2017JArSc..78...20O.
- ↑ "Origins, spread and ethnic association of European haplogroups and subclades". Eupedia. http://www.eupedia.com/europe/origins_haplogroups_europe.shtml#H.
- ↑ 28.0 28.1 Hay, Maciamo. "Haplogroup H (mtDNA)". http://www.eupedia.com/europe/Haplogroup_H_mtDNA.shtml. Access date 2015/10/02
- ↑ "Mitochondrial DNA haplogroups influence AIDS progression". AIDS 22 (18): 2429–39. November 2008. doi:10.1097/QAD.0b013e32831940bb. PMID 19005266.
- ↑ "The Haplogroup H&HV mtGenome Project: H3". mtDNA Test Results for Members. https://www.familytreedna.com/public/mtdna_h3/default.aspx?section=mtresults.
- ↑ 31.0 31.1 "Origin and expansion of haplogroup H, the dominant human mitochondrial DNA lineage in West Eurasia: the Near Eastern and Caucasian perspective". Molecular Biology and Evolution 24 (2): 436–48. February 2007. doi:10.1093/molbev/msl173. PMID 17099056.
- ↑ 32.0 32.1 32.2 "New population and phylogenetic features of the internal variation within mitochondrial DNA macro-haplogroup R0". PLOS ONE 4 (4): e5112. 2009. doi:10.1371/journal.pone.0005112. PMID 19340307. Bibcode: 2009PLoSO...4.5112A.
- ↑ "A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages". Nature Communications 4: 2543. 2013. doi:10.1038/ncomms3543. PMID 24104924. Bibcode: 2013NatCo...4.2543C.
- ↑ "Introducing the Algerian mitochondrial DNA and Y-chromosome profiles into the North African landscape". PLOS ONE 8 (2): e56775. 2013. doi:10.1371/journal.pone.0056775. PMID 23431392. Bibcode: 2013PLoSO...856775B.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. p. 41. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. p. 41. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. pp. 42–43. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ Dulias, Katharina; Birch, Steven; Wilson, James F.; Justeau, Pierre; Gandini, Francesca; Flaquer, Antònia; Soares, Pedro; Richards, Martin B. et al. (July 2019). "Maternal relationships within an Iron Age burial at the High Pasture Cave, Isle of Skye, Scotland" (in en). Journal of Archaeological Science 110: 104978. doi:10.1016/j.jas.2019.104978. Bibcode: 2019JArSc.110j4978D. https://linkinghub.elsevier.com/retrieve/pii/S0305440319300664.
- ↑ Template:GenBank
- ↑ Template:GenBank
- ↑ Template:GenBank
- ↑ Brook, Kevin Alan (Summer 2014). "The Genetics of Crimean Karaites". Karadeniz Araştırmaları (Journal of Black Sea Studies) 11 (42): 78–79. doi:10.12787/KARAM859. http://www.karamdergisi.com/Makaleler/909058854_5-%20Brook.pdf.
- ↑ Homo sapiens haplogroup H9a mitochondrion, complete genome GenBank: OQ981914.1.
- ↑ Matisoo-Smith, E.; Gosling, A. L.; Platt, D.; Kardailsky, O.; Prost, S.; Cameron-Christie, S.; Collins, C. J.; Boocock, J. et al. (2018). "Ancient mitogenomes of Phoenicians from Sardinia and Lebanon: A story of settlement, integration, and female mobility". PLOS ONE 13 (1): e0190169 in S1 Table. doi:10.1371/journal.pone.0190169. PMID 29320542. Bibcode: 2018PLoSO..1390169M.
- ↑ "A "Copernican" reassessment of the human mitochondrial DNA tree from its root". American Journal of Human Genetics 90 (4): 675–84. April 2012. doi:10.1016/j.ajhg.2012.03.002. PMID 22482806.
- ↑ Faustino de Carvalho, António (2014). Bom Santo cave (Lisbon) and the middle neolithic societies of southern Portugal. Faro: Universidade do Algarvede. ISBN 9789899766631. OCLC 946308166.
- ↑ Template:GenBank
- ↑ Template:GenBank
- ↑ Jones, Eppie R.; Gonzalez-Fortes, Gloria; Connell, Sarah; Siska, Veronika; Eriksson, Anders; Martiniano, Rui; McLaughlin, Russell L.; Gallego Llorente, Marcos et al. (16 November 2015). "Upper Palaeolithic genomes reveal deep roots of modern Eurasians". Nature Communications 6 (1): 8912. doi:10.1038/ncomms9912. PMID 26567969. Bibcode: 2015NatCo...6.8912J.
- ↑ Template:GenBank
- ↑ Template:GenBank
- ↑ 52.00 52.01 52.02 52.03 52.04 52.05 52.06 52.07 52.08 52.09 52.10 52.11 52.12 52.13 52.14 52.15 52.16 52.17 52.18 52.19 52.20 MtDNA Haplotree at Family Tree DNA
- ↑ Homo sapiens isolate 40_Mu mitochondrion, complete genome GenBank: MK491393.1.
- ↑ Homo sapiens isolate csct_007727 mitochondrion, complete genome GenBank: KY410191.1.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. p. 129. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ Homo sapiens isolate M943_H14a2_Roma mitochondrion, complete genome GenBank: KF055878.1.
- ↑ Homo sapiens haplogroup H14a2 mitochondrion, complete genome GenBank: MG786850.1.
- ↑ Homo sapiens isolate Assyrian_C188_H14b mitochondrion, complete genome GenBank: MK217254.1.
- ↑ Homo sapiens haplogroup H14b* mitochondrion, complete genome GenBank: KC911548.1.
- ↑ Homo sapiens haplogroup H14b mitochondrion, complete genome GenBank: KX784190.1.
- ↑ Homo sapiens isolate 33151 mitochondrion, complete genome GenBank: KY399186.1.
- ↑ Homo sapiens isolate Artsakh_69 haplogroup H14b3 mitochondrion, complete genome GenBank: MF362858.1.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. p. 135. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. p. 50. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ Homo sapiens haplogroup H16a mitochondrion, complete genome GenBank: MW538532.1.
- ↑ Homo sapiens isolate 3157345 mitochondrion, complete genome GenBank: JX153510.1.
- ↑ Homo sapiens isolate csct_007629 mitochondrion, complete genome GenBank: KY410158.1.
- ↑ Janis Kimsis, Elina Petersone-Gordina, Alise Poksane, Antonija Vilcāne, Joanna Moore, Guntis Gerhards, Renate Ranka (2023). "Application of natural sciences methodology in archaeological study of Iron Age burials in Latvia: pilot study"
- ↑ Homo sapiens isolate 1963 mitochondrion, complete genome GenBank: KY408165.1.
- ↑ 70.0 70.1 "Mitochondrial DNA haplogroup H structure in North Africa". BMC Genetics 10: 8. February 2009. doi:10.1186/1471-2156-10-8. PMID 19243582.
- ↑ Gómez-Carballa, Alberto; Pardo-Seco, Jacobo; Fachal, Laura; Vega, Ana; Cebey, Miriam; Martinón-Torres, Nazareth; Martinón-Torres, Federico; Salas, Antonio (15 October 2013). "Indian Signatures in the Westernmost Edge of the European Romani Diaspora: New Insight from Mitogenomes". PLOS ONE 8 (10): e75397. doi:10.1371/journal.pone.0075397. PMID 24143169. Bibcode: 2013PLoSO...875397G.
- ↑ (in en-US) Homo sapiens isolate S19_fi_ath haplogroup H45a mitochondrion, complete genome. 2019-11-10. https://www.ncbi.nlm.nih.gov/nuccore/MN516616.1.
- ↑ (in en-US) Homo sapiens haplogroup H45b mitochondrion, complete genome. 2018-01-24. https://www.ncbi.nlm.nih.gov/nuccore/MG807267.1.
- ↑ Brook, Kevin Alan (2022). The Maternal Genetic Lineages of Ashkenazic Jews. Academic Studies Press. pp. 53–54. doi:10.2307/j.ctv33mgbcn. ISBN 978-1644699843.
- ↑ (in en-US) Homo sapiens isolate ESP_19_00000014 mitochondrion, complete genome. 2020-05-27. https://www.ncbi.nlm.nih.gov/nuccore/MN046424.1.
- ↑ (in en-US) Homo sapiens isolate J189 mitochondrion, complete genome. 2018-02-14. https://www.ncbi.nlm.nih.gov/nuccore/MG646120.1.
- ↑ (in en-US) Homo sapiens isolate We1474 mitochondrion, complete genome. 2018-02-28. https://www.ncbi.nlm.nih.gov/nuccore/KU683584.1.
- ↑ Template:GenBank
- ↑ Template:GenBank
- ↑ (in en-US) Homo sapiens isolate We0454 mitochondrion, complete genome. 2018-02-28. https://www.ncbi.nlm.nih.gov/nuccore/KU683042.1.
- ↑ (in en-US) Homo sapiens isolate 23_Ps haplogroup H107 mitochondrion, complete genome. 2018-04-02. https://www.ncbi.nlm.nih.gov/nuccore/KY670896.1.
- ↑ "PhyloTree.org mtDNA subtree R0". https://phylotree.org/tree/R0.htm.
- ↑ "Mitochondrial haplogroups H and J: risk and protective factors for ischemic cardiomyopathy". PLOS ONE 7 (8): e44128. 2012. doi:10.1371/journal.pone.0044128. PMID 22937160. Bibcode: 2012PLoSO...744128F.
External links
- General
- Ian Logan's Mitochondrial DNA Site
- Mannis van Oven's Phylotree
- Haplogroup H
- mtDNA Haplogroup H Project at Family Tree DNA
- National Geographic's Spread of Haplogroup H, from National Geographic
- mtDNA Haplogroup H article at SNPedia
- Amelia's Helena
- "Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia". Molecular Biology and Evolution 21 (11): 2012–21. November 2004. doi:10.1093/molbev/msh209. PMID 15254257.
- Genebase's Tutorials on mtDNA Haplogroup H
- Genebase's Phylogenetic tree of mtDNA Haplogroup H
- Genebase's Geographical distribution of mtDNA Haplogroup H
- Haplogroup and Subcluster Frequencies for European Populations: "mtDna and the islands of the North Atlantic: estimating the proportions of Norse and Gaelic ancestry". American Journal of Human Genetics 68 (3): 723–37. March 2001. doi:10.1086/318785. PMID 11179019.
- Danish Demes Regional DNA Project: mtDNA Haplogroup H[yes|permanent dead link|dead link}}]
- Haplogroup H1
- Hope The H1 mtDNA Haplogroup Project
 |
