Biology:Genetic history of Africa
The genetic history of Africa is composed of the overall genetic history of African populations in Africa, including the regional genetic histories of North Africa, West Africa, East Africa, Central Africa, and Southern Africa, as well as the recent origin of modern humans in Africa. The Sahara served as a trans-regional passageway and place of dwelling for people in Africa during various humid phases[1][2][3] and periods throughout the history of Africa.[4][5]
Overview

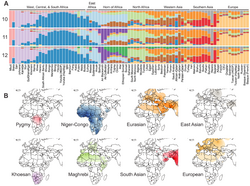
The peoples of Africa are characterized by regional genetic substructure and heterogeneity, depending on the respective ethno-linguistic identity, and, in part, explainable by the "multiregional evolution" of modern human lineages in various multiple regions of the African continent, as well as later admixture events, including back-migrations from Eurasia, of both highly differentiated West- and East-Eurasian components.[6]
Africans' genetic ancestry is largely partitioned by geography and language family, with populations belonging to the same ethno-linguistic groupings showing high genetic homogeneity and coherence. Gene flow, consistent with both short- and long-range migration events followed by extensive admixture and bottleneck events, have influenced the regional genetic makeup and demographic structure of Africans. The historical Bantu expansion had lasting impacts on the modern demographic make up of Africa, resulting in a greater genetic and linguistic homogenization.[7] Genetic, archeologic, and linguistic studies added extra insight into this movement: "Our results reveal a genetic continuum of Niger–Congo speaker populations across the continent and extend our current understanding of the routes, timing and extent of the Bantu migration."[8]
Overall, different African populations display genetic diversity and substructure, but can be clustered in distinct but partially overlapping groupings:[9]
- Khoisan hunter-gatherer lineages from Southern Africa represent the deepest lineages, forming a divergent and distinct cluster, shifted away from contemporary "Sub-Saharan Africans", and are further diverged from them than the various Eurasian lineages are. The diverging date of these "Southern hunter-gatherers" from all other human populations is estimated to over 100,000 years ago respectively, with the Khoisan later diverging into two subgroups, northern and southern Khoisan,~30,000 years ago.[lower-alpha 1]
- Rain forest foragers such as the Baka and the Mbuti diverged from other Sub-Saharan African groups over 60,000 years ago. Eastern groups such as the Mbuti split from Western groups such as the Baka ~20,000 years ago.
- The various Afroasiatic speakers are suggested to have diverged from other African groups ~50,000 years ago.[10]
- Niger-Congo and Nilo-Saharan speakers split around 28,000 years ago.[9]
- Austronesian-speaking Malagasy people in Madagascar have received significant East/Southeast Asian admixture, less among some groups of coastal Southern, Eastern and the Horn of Africa. The estimated date of geneflow is 2,200 years ago.[11][12][13][7][14][15]

Indigenous Africans

The Niger–Congo languages probably originated in or near the area where these languages were spoken prior to Bantu expansion (i.e. West Africa or Central Africa). Its expansion may have been associated with the expansion of agriculture, in the African Neolithic period, following the desiccation of the Sahara in c. 3500 BCE. Proto-Niger-Congo may have originated about 10,000 years before present in the "Green Sahara" of Africa (roughly the Sahel and southern Sahara), and that its dispersal can be correlated with the spread of the bow and arrow by migrating hunter-gatherers, which later developed agriculture.[17][18][19]
Although the validity of the Nilo-Saharan family remains controversial, the region between Chad, Sudan, and the Central African Republic is seen as a likely candidate for its homeland prior to its dispersal around 10,000–8,000 BCE.[20]
The Southern African hunter-gatherers (Khoisan) are suggested to represent the autochthonous hunter-gatherer population of southern Africa, prior to the expansion of Bantu-speakers from Western/Central Africa and East African pastoralists. Khoisan show evidence for Bantu-related admixture, ranging from nearly ~0% to up to ~87.1%.[21]
Out-of-Africa event


The "recent African origin of modern humans" model proposes a "single origin" of Homo sapiens in Africa the taxonomic sense. Recent genetic and archeologic data suggests that Homo sapiens-subgroups originated in multiple regions of Africa, not confined to a single region of origin. The H. sapiens ancestral to proper Eurasians most likely left Northeastern Africa between 50,000 and 100,000 years ago.[15] The "recent African origin" model proposes that all modern non-African populations descend from one or several waves of H. sapiens that left Africa 70,000-60,000 years ago.[22][23][24][25]

According to Durvasula et al. (2020), there are indications that 2% to 19% (≃6.6 to 7.0%) of the DNA of West African populations may have come from an unknown archaic hominin which split from the ancestor of humans and Neanderthals between 360 kya to 1.02 mya. However, Durvasula et al. (2020) also suggests that at least part of this archaic admixture is also present in Eurasians/non-Africans, and that the admixture event or events range from 0 to 124 ka B.P, which includes the period before the Out-of-Africa migration and prior to the African/Eurasian split (thus affecting in part the common ancestors of both Africans and Eurasians/non-Africans).[27][28][29] Chen et al. (2020) found that Africans have higher Neanderthal ancestry than previously thought. 2,504 African samples from all over Africa were analyzed and tested on Neanderthal ancestry. All African samples showed evidence for minor Neanderthal ancestry, but always at lower levels than observed in Eurasians.[30]
Geneflow between Eurasian and African populations

Significant Eurasian admixture is found in Northern Africa, and among specific ethnic groups of the Horn of Africa, as well as among the Malagasy people of Madagascar . Various genome studies found evidence for multiple prehistoric back-migrations from various Eurasian populations and subsequent admixture with native groups.[32] West-Eurasian geneflow arrived to Northern Africa during the Paleolithic (30,000 to 15,000 years ago), followed by other pre-Neolithic and Neolithic migration events. Genetic data on the Taforalt samples "demonstrated that Northern Africa received significant amounts of gene-flow from Eurasia predating the Holocene and development of farming practices". Medieval geneflow events, such as the Arab expansion also left traces in various African populations.[15][31][33] Pickrell et al. (2014) indicated that Western Eurasian ancestry eventually arrived through Northeast Africa (particularly the Horn of Africa) to Southern Africa.[34]
Ramsay et al. (2018) also found evidence for significant Western Eurasian admixture in various parts of Africa, from both ancient and more recent migrations, being highest among populations from Northern Africa, and some groups of the Horn of Africa:[35]
In addition to the intrinsic diversity within the continent due to population structure and isolation, migration of Eurasian populations into Africa has emerged as a critical contributor to the genetic diversity. These migrations involved the influx of different Eurasian populations at different times and to different parts of Africa. Comprehensive characterization of the details of these migrations through genetic studies on existing populations could help to explain the strong genetic differences between some geographically neighbouring populations.
This distinctive Eurasian admixture appears to have occurred over at least three time periods with ancient admixture in central west Africa (e.g., Yoruba from Nigeria) occurring between ~7.5 and 10.5 kya, older admixture in east Africa (e.g., Ethiopia) occurring between ~2.4 and 3.2 kya and more recent admixture between ~0.15 and 1.5 kya in some east African (e.g., Kenyan) populations.
Subsequent studies based on LD decay and haplotype sharing in an extensive set of African and Eurasian populations confirmed the presence of Eurasian signatures in west, east and southern Africans. In the west, in addition to Niger-Congo speakers from The Gambia and Mali, the Mossi from Burkina Faso showed the oldest Eurasian admixture event ~7 kya. In the east, these analyses inferred Eurasian admixture within the last 4000 years in Kenya.[35]
There is no definitive agreement on when or where the original homeland of the Afroasiatic language family existed. Some have suggested that they were spread by people with largely West-Eurasian ancestry during the Neolithic Revolution, towards Northern Africa and the Horn of Africa, outgoing from the Middle East, specifically from the Levant. This hypothesis does not account for the domestication of plants endemic to the Horn of Africa such as teff, ensete, and Niger seed, nor does it account for the lack of evidence of intrusive agricultural populations or the cultivation of wheat, barley, or sorghum in that region prior to 3000 B.C.[36][37] Others argue that the first speakers of Proto-Afroasiatic were based in Northeast Africa because that region includes the majority of the diversity of the Afroasiatic language family and has very diverse groups in close geographic proximity, which is sometimes considered a telltale sign for a linguistic geographic origin.[38] A subset of the Proto-Afroasiatic population would have migrated to the Levant during the late Paleolithic, merging with local West-Eurasians and resulting in a population which would later give rise to Natufian culture, associated with the early development of agriculture and early Afroasiatic languages, or specifically pre-proto-Semitic.[39][40][page needed][41][42][43][44] In addition, Y-haplogroup sub-lineage E-M215 (also known as "E1b1b) and its derivative E-M35 are quite common among Afroasiatic speakers, and southwestern Ethiopia is a plausible source of these haplogroups.[45] The linguistic group and carriers of this lineage would have arisen and dispersed together from Northeast Africa in the Mesolithic, plausibly having already developed subsistence patterns of pastoralism and intensive plant usage and collection.[46][47][48][49] According to historian and linguist Christopher Ehret, the form of intensive plant collection practiced by the Proto-Afroasiatic population in Northeast Africa may have been a precursor to the agricultural practices that would later independently develop in the Fertile Crescent and the Horn of Africa.[43][50][51]
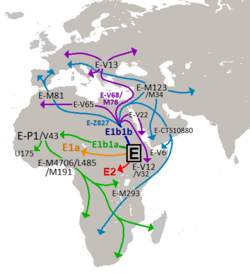
Horn of Africa
While many studies conducted on Horn of Africa populations estimate a West-Eurasian admixture event around 3,000 years ago,[52][35][34][53] Hodgson et al. (2014) found a distinct West-Eurasian ancestral component among studied Afroasiatic-speaking groups in the Horn of Africa (and to a lesser extent in North Africa and West Asia), most prevalent among the Somali. This ancestral component dubbed "Ethio-Somali" is most closely related to the "Maghrebi" (peaking in Tunisians) component and is believed to have diverged from other non-African ancestries around 23,000 years ago, and migrated back to Africa prior to developing agriculture (12–23 ka) from the Near East. This population would have crossed via the Sinai Peninsula and then split into two, with one branch continuing west across North Africa and the other heading south into the Horn of Africa. The authors propose that "Ethio-Somali" may have been a substantial ancestral component of the Proto-Afroasiatic-speaking population. Later migration from Arabia into the HOA beginning around 3 ka would explain the origin of the Ethiosemitic languages at this time.[41] An mtDNA analysis by Gandini et al. (2016) has produced additional evidence in support of a pre-agricultural back-migration from West-Eurasia into the Horn of Africa with an estimated date of arrival into the Horn of Africa in the early Holocene, possibly as a result of obsidian exchange networks across the Red Sea.[54] Hodgson et al. also confirmed the existence of an ancestral component indigenous to the Horn of Africa - "Ethiopic" or "Omotic" (Pagani et al.) - which is most prevalent among speakers of the Omotic branch of Afroasiatic in southwestern Ethiopia.[41][52] This lineage is associated with that of a 4,500 year-old fossil (Mota) found in a cave in southwestern Ethiopia, which has high genetic affinity to modern Ethiopian groups, especially the endogamous blacksmith caste of the Omotic Aari people. Like Mota, Aari blacksmiths do not show evidence for admixture with West-Eurasians, demonstrating a degree of population continuity in this region for at least 4,500 years. In a comparative analysis of Mota's genome referencing modern populations, Gallego et al. (2016) concluded that the divergence of Omotic from other Afroasiatic languages may have resulted from the relative isolation of its speakers from external groups.[55] In an analysis of 68 Ethiopian ethnic groups, Lopez et al. (2021) revealed that several groups belonging to the three AA classifications of Cushitic, Omotic and Semitic show high genetic similarity to each other on average. Furthermore, the Nilo-Saharan speakers in the southwest shared more recent ancestry with Bantu and Nilotics, in contrast Afro-Asiatic speakers in the northeast shared more recent ancestry with Egyptians and other West Eurasians. The data also supported widespread recent intermixing among various ethnic groups.[56]
Madagascar

Specific East Asian-related ancestry is found among the Malagasy speakers of Madagascar at a medium frequency. The presence of this East Asian-related ancestry is mostly linked to the Austronesian peoples expansion from Southeast Asia.[57][58][59][60] The peoples of Borneo were identified to resemble the East Asian voyagers, who arrived on Madagascar. East Asian ancestry among Malagasy people was estimated at a mean average of 33%, but as high as ~75% among some Highlander groups and upper caste groups.[61][62][60]
Northern Africa
Dobon et al. (2015) identified an autosomal ancestral component that is commonly found among modern Afroasiatic-speaking populations (as well as Nubians) in Northeast Africa. This Coptic component peaks among Copts in Sudan, which is differentiated by its lack of Arab influence, but shares common ancestry with the North African/Middle Eastern populations. It appears alongside a component that defines Nilo-Saharan speakers of southwestern Sudan and South Sudan.[63] Arauna et al. (2017), analyzing existing genetic data obtained from Northern African populations, such as Berbers, described them as a mosaic of North African (Taforalt), Middle Eastern, European (Early European Farmers), and Sub-Saharan African-related ancestries.[64]
Chen et al. (2020) analyzed 2,504 African samples from all over Africa, and found archaic Neanderthal ancestry, among all tested African samples at low frequency. They also identified a European-related (West-Eurasian) ancestry segment, which seems to largely correspond with the detected Neanderthal ancestry components. European-related admixture among Africans was estimated to be between ~0% to up to ~30%, with a peak among Northern Africans.[14] According to Chen et al. (2020), "These data are consistent with the hypothesis that back-migration contributed to the signal of Neanderthal ancestry in Africans. Furthermore, the data indicates that this back-migration came after the split of Europeans and East Asians, from a population related to the European lineage."[14]
There is a minor geneflow from North Africa in parts of Southern Europe, this is supported by the presence of an African-specific mitochondrial haplogroup among one of four 4,000 year old samples.[65] Multiple studies found also evidence for geneflow of African ancestry towards Eurasia, specifically Europe and the Middle East. The analysis of 40 different West-Eurasian populations found African admixture at a frequency of 0% to up to ~15%.[66][67][68][69]
Western Africa
Hollfelder et al. (2021) concluded that West African Yoruba people, which were previously used as "unadmixed reference population" for indigenous Africans, harbor minor levels of Neanderthal ancestry, which can be largely associated with back-migration of an "Ancestral European-like" source population.[6]
A genome-wide study of a Fulani community from Burkina Faso inferred two major admixture events in this group, dating to ~1800 ya, and 300 ya. The first admixture event took place between the West African ancestors of the Fula and ancestral North African nomadic groups. The second admixture event, relatively recent, inferred a source from Southwestern Europe, or suggests either an additional gene flow between the Fulani and Northern African groups, who carry admixture proportions from Europeans.[70] Sahelian populations like the Toubou also showed admixture coming from Eurasians.[71]
Southern Africa
Low levels of West Eurasian ancestry (European or Middle Eastern) are found in Khoe–Kwadi Khoesan-speakers. It could have been acquired indirectly by admixture with migrating pastoralists from East Africa. This hypothesis of gene flow from eastern to southern Africa is further supported by other genetic and archaeological data documenting the spread of pastoralism from East to South Africa.[72]
Regional genomic overview
North Africa
Archaic Human DNA
While Denisovan and Neanderthal ancestry in non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.[73]
Ancient DNA
Daniel Shriner (2018), using modern populations as a reference, showed that the Natufians carried 61.2% Arabian, 21.2% Northern African, 10.9% Western Asian, and a small portion of Eastern African ancestry at 6.8%, which is associated with the modern Omotic-speaking groups found in southern Ethiopia.[45]
Egypt
Khnum-aa, Khnum-Nakht, Nakht-Ankh and JK2911 carried maternal haplogroup M1a1.[74][52]
Djehutynakht (10A) carried maternal haplogroup U5b2b5.[75] JK2888 carried maternal haplogroup U6a2.[52]
Thuya, Tiye, Tutankhamen's mother, and Tutankhamen carried the maternal haplogroup K.[74]
JK2134 carried maternal haplogroup J1d[52] and JK2887 carried maternal haplogroup J2a1a1.[52]
Amenhotep III, Akhenaten, and Tutankhamen carried the paternal haplogroup R1b.[74]
Ramesses III and "Unknown Man E", possibly Pentawere, carried paternal haplogroup E1b1a.[74][76][77]
JK2134 and JK2911 carried paternal haplogroup J.[52]
Takabuti carried maternal haplogroup H4a1[78] and YM:KMM A 63 carried maternal haplogroup HV.[79]
OM:KMM A 64 carried maternal haplogroup T2c1a.[79]
JK2888 carried paternal haplogroup E1b1b1a1b2.[52]
Libya
At Takarkori rockshelter, in Libya, two naturally mummified women, dated to the Middle Pastoral Period (7000 BP), carried basal maternal haplogroup N.[80]
Morocco
Van de Loorsdrecht et al. (2018) found that of seven samples of Taforalts of Morocco, radiocarbon dated to between 15,100 cal BP and 13,900 cal BP, six were found to carry maternal haplogroup U6a, and one was found to carry maternal haplogroup M1b. Six of six males were found to carry paternal haplogroup E1b1b. They were found to harbor 63.5% Natufian-related ancestry and 36.5% Sub-Saharan African-related ancestry. The Sub-Saharan component is most strongly drawn out by modern West African groups such as the Yoruba and the Mende. The samples also contain an additional affinity to South, Central, and East African outgroups that cannot be explained by any known ancient or modern populations.[81] When projected onto a principal component analysis graph of African and west Eurasian populations, the Taforalt individuals form a distinct cluster in an intermediate position between modern North Africans (e.g., Berbers, Mozabites, Saharawis) and East Africans (e.g., Afars, Oromos, Somalis).[81] Jeong (2020), comparing the Taforalt people of the Iberomaurusian culture to modern populations, found that the Taforalt's Sub-Saharan African genetic component may be best represented by modern West Africans (e.g., Yoruba).[82]
Y-Chromosomal DNA
Mitochondrial DNA
Amid the Holocene, including the Holocene Climate Optimum in 8000 BP, Africans bearing haplogroup L2 spread within West Africa and Africans bearing haplogroup L3 spread within East Africa.[83] As the largest migration since the Out of Africa migration, migration from Sub-Saharan Africa toward the North Africa occurred, by West Africans, Central Africans, and East Africans, resulting in migrations into Europe and Asia; consequently, Sub-Saharan African mitochondrial DNA was introduced into Europe and Asia.[83] During the early period of the Holocene, 50% of Sub-Saharan African mitochondrial DNA was introduced into North Africa by West Africans and the other 50% was introduced by East Africans.[83] During the modern period, a greater number of West Africans introduced Sub-Saharan African mitochondrial DNA into North Africa than East Africans.[83]
Mitochondrial haplogroups L3, M, and N are found among Sudanese peoples (e.g., Beja, Nilotics, Nuba, Nubians), who have no known interaction (e.g., history of migration/admixture) with Europeans or Asians; rather than having developed in a post-Out-of-Africa migration context, mitochondrial macrohaplogroup L3/M/N and its subsequent development into distinct mitochondrial haplogroups (e.g., Haplogroup L3, Haplogroup M, Haplogroup N) may have occurred in East Africa at a time that considerably predates the Out-of-Africa migration event of 50,000 BP.[84]
Autosomal DNA
Medical DNA
The genomes of Africans commonly found to undergo adaptation are regulatory DNA, and many cases of adaptation found among Africans relate to diet, physiology, and evolutionary pressures from pathogens.[85]
Lactase Persistence
Neolithic agriculturalists, who may have resided in Northeast Africa and the Near East, may have been the source population for lactase persistence variants, including –13910*T, and may have been subsequently supplanted by later migrations of peoples.[86] The Sub-Saharan West African Fulani, the North African Tuareg, and European agriculturalists, who are descendants of these Neolithic agriculturalists, share the lactase persistence variant –13910*T.[86] While shared by Fulani and Tuareg herders, compared to the Tuareg variant, the Fulani variant of –13910*T has undergone a longer period of haplotype differentiation.[86] The Fulani lactase persistence variant –13910*T may have spread, along with cattle pastoralism, between 9686 BP and 7534 BP, possibly around 8500 BP; corroborating this timeframe for the Fulani, by at least 7500 BP, there is evidence of herders engaging in the act of milking in the Central Sahara.[86]
West Africa
Archaic Human DNA
Archaic traits found in human fossils of West Africa (e.g., Iho Eleru fossils, which dates to 13,000 BP) and Central Africa (e.g., Ishango fossils, which dates between 25,000 BP and 20,000 BP) may have developed as a result of admixture between archaic humans and modern humans or may be evidence of late-persisting early modern humans.[73] While Denisovan and Neanderthal ancestry in non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.[73]
Ancient DNA
As of 2017, human ancient DNA has not been found in the region of West Africa.[87] As of 2020, human ancient DNA has not been forthcoming in the region of West Africa.[82]
Y-Chromosomal DNA
Eight male individuals from Guinea Bissau, two male individuals from Niger, one male individual from Mali, and one male individual from Cabo Verde carried haplogroup A1a.[88]
As a result of haplogroup D0, a basal branch of haplogroup DE, being found in three Nigerian men, it may be the case that haplogroup DE, as well as its sublineages D0 and E, originated in Africa.[89]
As of 19,000 years ago, Africans, bearing haplogroup E1b1a-V38, likely traversed across the Sahara, from east to west.[90] E1b1a1-M2 likely originated in West Africa or Central Africa.[91]
Mitochondrial DNA
Around 18,000 BP, Mende people, along with Gambian peoples, grew in population size.[92]
In 15,000 BP, Niger-Congo speakers may have migrated from the Sahelian region of West Africa, along the Senegal River, and introduced L2a1 into North Africa, resulting in modern Mauritanian peoples and Berbers of Tunisia inheriting it.[93]
Between 11,000 BP and 10,000 BP, Yoruba people and Esan people grew in population size.[92]
As early as 11,000 years ago, Sub-Saharan West Africans, bearing macrohaplogroup L (e.g., L1b1a11, L1b1a6a, L1b1a8, L1b1a9a1, L2a1k, L3d1b1a), may have migrated through North Africa and into Europe, mostly into southern Europe (e.g., Iberia).[94]
Autosomal DNA
During the early period of the Holocene, in 9000 BP, Khoisan-related peoples admixed with the ancestors of the Igbo people, possibly in the western Sahara.[95][96]
Between 2000 BP and 1500 BP, Nilo-Saharan-speakers may have migrated across the Sahel, from East Africa into West Africa, and admixed with Niger-Congo-speaking Berom people.[97] In 710 CE, West African-related populations (e.g., Niger-Congo-speaking Berom people, Bantu-speakers) and East African-related populations (Nilo-Saharan-speaking Ethiopians, Nilo-Saharan-speaking Chadians) admixed with one another in northern Nigeria and northern Cameroon.[98]
Fan et al. (2019) found that the Fulani people show genetic affinity to isolated Afroasiatic-speaking groups in Eastern Africa, specifically Omotic-speakers such as the Aari people. While the Fulani have nearly exclusive indigenous African ancestry (defined by West and East African ancestry), they also show traces of West-Eurasian-like admixture, supporting an ancestral homeland somewhere in North or Eastern Africa, and westwards expansion during the Neolithic, possibly caused by the arrival and expansion of West-Eurasian-related groups.[99] Fan et al. (2023) found that the Fulani, who have 50% Amhara-related and 50% Tikari-related ancestry as well as occupy regions such as West Africa, Central Africa, and the Sudan as nomadic herders, may have initially been Afroasiatic speakers that subsequently underwent language replacement and became Niger-Congo speakers.[100]
Medical DNA
The genomes of Africans commonly found to undergo adaptation are regulatory DNA, and many cases of adaptation found among Africans relate to diet, physiology, and evolutionary pressures from pathogens.[85] Throughout Sub-Saharan Africa, genetic adaptation (e.g., rs334 mutation, Duffy blood group, increased rates of G6PD deficiency, sickle cell disease) to malaria has been found among Sub-Saharan Africans, which may have initially developed in 7300 BP.[85] Sub-Saharan Africans have more than 90% of the Duffy-null genotype.[101]
Pediculus
During the Copper Age and early Islamic era of ancient Israel, West Africans may have migrated into ancient Israel and introduced head louse from West Africa.[102]
Sickle Cell
Amid the Green Sahara, the mutation for sickle cell originated in the Sahara[90] or in the northwest forest region of western Central Africa (e.g., Cameroon)[90][103] by at least 7,300 years ago,[90][103] though possibly as early as 22,000 years ago.[104][103] The ancestral sickle cell haplotype to modern haplotypes (e.g., Cameroon/Central African Republic and Benin/Senegal haplotypes) may have first arose in the ancestors of modern West Africans, bearing haplogroups E1b1a1-L485 and E1b1a1-U175 or their ancestral haplogroup E1b1a1-M4732.[90] West Africans (e.g., Yoruba and Esan of Nigeria), bearing the Benin sickle cell haplotype, may have migrated through the northeastern region of Africa into the western region of Arabia.[90] West Africans (e.g., Mende of Sierra Leone), bearing the Senegal sickle cell haplotype,[105][90] may have migrated into Mauritania (77% modern rate of occurrence) and Senegal (100%); they may also have migrated across the Sahara, into North Africa, and from North Africa, into Southern Europe, Turkey, and a region near northern Iraq and southern Turkey.[105] Some may have migrated into and introduced the Senegal and Benin sickle cell haplotypes into Basra, Iraq, where both occur equally.[105] West Africans, bearing the Benin sickle cell haplotype, may have migrated into the northern region of Iraq (69.5%), Jordan (80%), Lebanon (73%), Oman (52.1%), and Egypt (80.8%).[105]
Schistosomes
According to Steverding (2020), while not definite: Near the African Great Lakes, schistosomes (e.g., S. mansoni, S. haematobium) underwent evolution.[106] Subsequently, there was an expansion alongside the Nile.[106] From Egypt, the presence of schistosomes may have expanded, via migratory Yoruba people, into Western Africa.[106] Thereafter, schistosomes may have expanded, via migratory Bantu peoples, into the rest of Sub-Saharan Africa (e.g., Southern Africa, Central Africa).[106]
Thalassemia
Through pathways taken by caravans, or via travel amid the Almovarid period, a population (e.g., Sub-Saharan West Africans) may have introduced the –29 (A → G) β-thalassemia mutation (found in notable amounts among African-Americans) into the North African region of Morocco.[107]
Domesticated Animal DNA
While the Niger-Congo migration may have been from West Africa into Kordofan, possibly from Kordofan, Sudan, Niger-Congo speakers accompanied by undomesticated helmeted guineafowls, may have traversed into West Africa, domesticated the helmeted guineafowls by 3000 BCE, and via the Bantu expansion, traversed into other parts of Sub-Saharan Africa (e.g., Central Africa, East Africa, Southern Africa).[108]
Central Africa
Archaic Human DNA
Archaic traits found in human fossils of West Africa (e.g., Iho Eleru fossils, which dates to 13,000 BP) and Central Africa (e.g., Ishango fossils, which dates between 25,000 BP and 20,000 BP) may have developed as a result of admixture between archaic humans and modern humans or may be evidence of late-persisting early modern humans.[73] While Denisovan and Neanderthal ancestry in non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.[73]
Ancient DNA
In 4000 BP, there may have been a population that traversed from Africa (e.g., West Africa or West-Central Africa), through the Strait of Gibraltar, into the Iberian peninsula, where admixing between Africans and Iberians (e.g., of northern Portugal, of southern Spain ) occurred.[109]
Cameroon
West African hunter-gatherers, in the region of western Central Africa (e.g., Shum Laka, Cameroon), particularly between 8000 BP and 3000 BP, were found to be related to modern Central African hunter-gatherers (e.g., Baka, Bakola, Biaka, Bedzan).[110]
Democratic Republic of Congo
At Kindoki, in the Democratic Republic of Congo, there were three individuals, dated to the protohistoric period (230 BP, 150 BP, 230 BP); one carried haplogroups E1b1a1a1d1a2 (E-CTS99, E-CTS99) and L1c3a1b, another carried haplogroup E (E-M96, E-PF1620), and the last carried haplogroups R1b1 (R-P25 1, R-M415) and L0a1b1a1.[111][112]
Y-Chromosomal DNA
Haplogroup R-V88 may have originated in western Central Africa (e.g., Equatorial Guinea), and, in the middle of the Holocene, arrived in North Africa through population migration.[113]
Mitochondrial DNA
In 150,000 BP, Africans (e.g., Central Africans, East Africans) bearing haplogroup L1 diverged.[83] Between 75,000 BP and 60,000 BP, Africans bearing haplogroup L3 emerged in East Africa and eventually migrated into and became present in modern West Africans, Central Africans, and non-Africans.[83] Amid the Holocene, including the Holocene Climate Optimum in 8000 BP, Africans bearing haplogroup L2 spread within West Africa and Africans bearing haplogroup L3 spread within East Africa.[83] As the largest migration since the Out of Africa migration, migration from Sub-Saharan Africa toward the North Africa occurred, by West Africans, Central Africans, and East Africans, resulting in migrations into Europe and Asia; consequently, Sub-Saharan African mitochondrial DNA was introduced into Europe and Asia.[83]
Mitochondrial haplogroup L1c is strongly associated with pygmies, especially with Bambenga groups.[114] L1c prevalence was variously reported as: 100% in Ba-Kola, 97% in Aka (Ba-Benzélé), and 77% in Biaka,[115] 100% of the Bedzan (Tikar), 97% and 100% in the Baka people of Gabon and Cameroon, respectively,[116] 97% in Bakoya (97%), and 82% in Ba-Bongo.[114] Mitochondrial haplogroups L2a and L0a are prevalent among the Bambuti.[114][117]
Autosomal DNA
Genetically, African pygmies have some key difference between them and Bantu peoples.[118][119]
Medical DNA
Evidence suggests that, when compared to other Sub-Saharan African populations, African pygmy populations display unusually low levels of expression of the genes encoding for human growth hormone and its receptor associated with low serum levels of insulin-like growth factor-1 and short stature.[120]
The genomes of Africans commonly found to undergo adaptation are regulatory DNA, and many cases of adaptation found among Africans relate to diet, physiology, and evolutionary pressures from pathogens.[85] Throughout Sub-Saharan Africa, genetic adaptation (e.g., rs334 mutation, Duffy blood group, increased rates of G6PD deficiency, sickle cell disease) to malaria has been found among Sub-Saharan Africans, which may have initially developed in 7300 BP.[85] Sub-Saharan Africans have more than 90% of the Duffy-null genotype.[101] In the rainforests of Central Africa, genetic adaptation for non-height-related factors (e.g., immune traits, reproduction, thyroid function) and short stature (e.g., EHB1 and PRDM5 – bone synthesis; OBSCN and COX10 – muscular development; HESX1 and ASB14 – pituitary gland’s growth hormone production/secretion) has been found among rainforest hunter-gatherers.[85]
Eastern Africa
From the region of Kenya and Tanzania to South Africa , eastern Bantu-speaking Africans constitute a north to south genetic cline; additionally, from eastern Africa to toward southern Africa, evidence of genetic homogeneity is indicative of a serial founder effect and admixture events having occurred between Bantu-speaking Africans and other African populations by the time the Bantu migration had spanned into South Africa.[85]
Archaic Human DNA
While Denisovan and Neanderthal ancestry in non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.[73]
Ancient DNA
Ethiopia
At Mota, in Ethiopia, an individual, estimated to date to the 5th millennium BP, carried haplogroups E1b1 and L3x2a.[121][122] The individual of Mota is genetically related to groups residing near the region of Mota, and in particular, are considerably genetically related to the Aari people, especially the blacksmith caste of that group.[123][124]
Kenya
At Jawuoyo Rockshelter, in Kisumu County, Kenya, a forager of the Later Stone Age carried haplogroups E1b1b1a1b2/E-V22 and L4b2a2c.[125][126]
At Ol Kalou, in Nyandarua County, Kenya, a pastoralist of the Pastoral Neolithic carried haplogroups E1b1b1b2b2a1/E-M293 and L3d1d.[125][126]
At Kokurmatakore, in Marsabit County, Kenya, a pastoralist of the Pastoral Iron Age carried haplogroups E1b1b1/E-M35 and L3a2a.[125][126]
At White Rock Point, in Homa Bay County, Kenya, there were two foragers of the Later Stone Age; one carried haplogroups BT (xCT), likely B, and L2a4, and another probably carried haplogroup L0a2.[125][126]
At Nyarindi Rockshelter, in Kenya, there were two individuals, dated to the Later Stone Age (3500 BP); one carried haplogroup L4b2a and another carried haplogroup E (E-M96, E-P162).[111][112]
At Lukenya Hill, in Kenya, there were two individuals, dated to the Pastoral Neolithic (3500 BP); one carried haplogroups E1b1b1b2b (E-M293, E-CTS10880) and L4b2a2b, and another carried haplogroup L0f1.[111][112]
At Hyrax Hill, in Kenya, an individual, dated to the Pastoral Neolithic (2300 BP), carried haplogroups E1b1b1b2b (E-M293, E-M293) and L5a1b.[111][112]
At Molo Cave, in Kenya, there were two individuals, dated to the Pastoral Neolithic (1500 BP); while one had haplogroups that went undetermined, another carried haplogroups E1b1b1b2b (E-M293, E-M293) and L3h1a2a1.[111][112]
At Kakapel, in Kenya, there were three individuals, one dated to the Later Stone Age (3900 BP) and two dated to the Later Iron Age (300 BP, 900 BP); one carried haplogroups CT (CT-M168, CT-M5695) and L3i1, another carried haplogroup L2a1f, and the last carried haplogroup L2a5.[111][112]
At Panga ya Saidi, in Kenya, an individual, estimated to date between 496 BP and 322 BP, carried haplogroups E1b1b1b2 and L4b2a2.[127]
At Kilifi, Mtwapa, in Kenya, an individual, dated between 1250 CE and 1650 CE, carried haplogroup L3b1a1a.[128]
Tanzania
At Mlambalasi rockshelter, in Tanzania, an individual, dated between 20,345 BP and 17,025 BP, carried undetermined haplogroups.[129]
At Gishimangeda Cave, in Karatu District, Tanzania, there were eleven pastoralists of the Pastoral Neolithic; one carried haplogroups E1b1b1a1b2/E-V22 and HV1b1, another carried haplogroup L0a, another carried haplogroup L3x1, another carried haplogroup L4b2a2b, another carried haplogroups E1b1b1b2b2a1/E-M293 and L3i2, another carried haplogroup L3h1a2a1, another carried haplogroups E1b1b1b2b2/E-V1486, likely E-M293 and L0f2a1, and another carried haplogroups E1b1b1b2b2/E-V1486, likely E-M293, and T2+150; while most of the haplogroups among three pastoralists went undetermined, one was determined to carry haplogroup BT, likely B.[125][126]
At Kilwa, Coast, in Tanzania, an individual, dated between 1300 CE and 1600 CE, carried haplogroups J2a2a1a1a2a~ and L2a1h.[128]
At Lindi, in Tanzania, an individual, dated between 1511 cal CE and 1664 cal CE, carried haplogroups E1b1a1a1a2a1a3a1d~ and L0a1a2.[128]
At Makangale Cave, on Pemba Island, Tanzania, an individual, estimated to date between 1421 BP and 1307 BP, carried haplogroup L0a.[127]
At Songo Mnara, in Tanzania, an individual, dated between 1294 cal CE and 1392 cal CE, carried haplogroups R1a and L3e3a.[128]
Uganda
At Munsa, in Uganda, an individual, dated to the Later Iron Age (500 BP), carried haplogroup L3b1a1.[111][112]
Y-Chromosomal DNA
As of 19,000 years ago, Africans, bearing haplogroup E1b1a-V38, likely traversed across the Sahara, from east to west.[90]
Before the slave trade period, East Africans, who carried haplogroup E1b1a-M2, expanded into Arabia, resulting in various rates of inheritance throughout Arabia (e.g., 2.8% Qatar, 3.2% Yemen, 5.5% United Arab Emirates, 7.4% Oman).[130]
Mitochondrial DNA
In 150,000 BP, Africans (e.g., Central Africans, East Africans) bearing haplogroup L1 diverged.[83] In 130,000 BP, Africans bearing haplogroup L5 diverged in East Africa.[83] Between 130,000 BP and 75,000 BP, behavioral modernity emerged among Southern Africans and long-term interactions between the regions of Southern Africa and Eastern Africa became established.[83] Between 75,000 BP and 60,000 BP, Africans bearing haplogroup L3 emerged in East Africa and eventually migrated into and became present in modern West Africans, Central Africans, and non-Africans.[83] Amid the Holocene, including the Holocene Climate Optimum in 8000 BP, Africans bearing haplogroup L2 spread within West Africa and Africans bearing haplogroup L3 spread within East Africa.[83] As the largest migration since the Out of Africa migration, migration from Sub-Saharan Africa toward the North Africa occurred, by West Africans, Central Africans, and East Africans, resulting in migrations into Europe and Asia; consequently, Sub-Saharan African mitochondrial DNA was introduced into Europe and Asia.[83] During the early period of the Holocene, 50% of Sub-Saharan African mitochondrial DNA was introduced into North Africa by West Africans and the other 50% was introduced by East Africans.[83] During the modern period, a greater number of West Africans introduced Sub-Saharan African mitochondrial DNA into North Africa than East Africans.[83] Between 15,000 BP and 7000 BP, 86% of Sub-Saharan African mitochondrial DNA was introduced into Southwest Asia by East Africans, largely in the region of Arabia, which constitute 50% of Sub-Saharan African mitochondrial DNA in modern Southwest Asia.[83] In the modern period, 68% of Sub-Saharan African mitochondrial DNA was introduced by East Africans and 22% was introduced by West Africans, which constitutes 50% of Sub-Saharan African mitochondrial DNA in modern Southwest Asia.[83]
Autosomal DNA
Across all areas of Madagascar , the average ancestry for the Malagasy people was found to be 4% West Eurasian, 37% Austronesian, and 59% Bantu.[61]
Medical DNA
The genomes of Africans commonly found to undergo adaptation are regulatory DNA, and many cases of adaptation found among Africans relate to diet, physiology, and evolutionary pressures from pathogens.[85] Throughout Sub-Saharan Africa, genetic adaptation (e.g., rs334 mutation, Duffy blood group, increased rates of G6PD deficiency, sickle cell disease) to malaria has been found among Sub-Saharan Africans, which may have initially developed in 7300 BP.[85] Sub-Saharan Africans have more than 90% of the Duffy-null genotype.[101] In the highlands of Ethiopia, genetic adaptation (e.g., rs10803083, an SNP associated with the rate and function of hemoglobin; BHLHE41, a gene associated with circadian rhythm and hypoxia response; EGNL1, a gene strongly associated with oxygen homeostasis in mammals) to hypoxia and low atmospheric pressure has been found among the Amhara people, which may have developed within the past 5000 years.[85] In Tanzania, genetic adaptation (e.g., greater amount of amylase genes than in African populations that consume low-starch foods) has been found in the Hadza people due to a food diet that especially includes consumption of tubers.[85]
Southern Africa
From the region of Kenya and Tanzania to South Africa , eastern Bantu-speaking Africans constitute a north to south genetic cline; additionally, from eastern Africa to toward southern Africa, evidence of genetic homogeneity is indicative of a serial founder effect and admixture events having occurred between Bantu-speaking Africans and other African populations by the time the Bantu migration had spanned into South Africa.[85]
Archaic Human DNA
While Denisovan and Neanderthal ancestry in non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.[73]
Ancient DNA
Three Later Stone Age hunter-gatherers carried ancient DNA similar to Khoisan-speaking hunter-gatherers.[131] Prior to the Bantu migration into the region, as evidenced by ancient DNA from Botswana, East African herders migrated into Southern Africa.[131] Out of four Iron Age Bantu agriculturalists of West African origin, two earlier agriculturalists carried ancient DNA similar to Tsonga and Venda peoples and the two later agriculturalists carried ancient DNA similar to Nguni people; this indicates that there were various movements of peoples in the overall Bantu migration, which resulted in increased interaction and admixing between Bantu-speaking peoples and Khoisan-speaking peoples.[131]
Botswana
At Nqoma, in Botswana, an individual, dated to the Early Iron Age (900 BP), carried haplogroup L2a1f.[111][112]
At Taukome, in Botswana, an individual, dated to the Early Iron Age (1100 BP), carried haplogroups E1b1a1 (E-M2, E-Z1123) and L0d3b1.[111][112]
At Xaro, in Botswana, there were two individuals, dated to the Early Iron Age (1400 BP); one carried haplogroups E1b1a1a1c1a and L3e1a2, and another carried haplogroups E1b1b1b2b (E-M293, E-CTS10880) and L0k1a2.[111][112]
Malawi
At Fingira rockshelter, in Malawi, an individual, dated between 6179 BP and 2341 BP, carried haplogroups B2 and L0d1.[129]
At Chencherere, in Malawi, an individual, estimated to date between 5400 BP and 4800 BP, carried haplogroup L0k2.[127]
At Hora 1 rockshelter, in Malawi, an individual, dated between 16,897 BP and 15,827 BP, carried haplogroups B2b and L5b.[129]
South Africa
At Doonside, in South Africa , an individual, estimated to date between 2296 BP and 1910 BP, carried haplogroup L0d2.[132][133]
At Ballito Bay, South Africa , an individual, estimated to date between 1986 BP and 1831 BP, carried haplogroups A1b1b2 and L0d2c1.[132][133]
At Kalemba rockshelter, in Zambia, an individual, dated between 5285 BP and 4975 BP, carried haplogroup L0d1b2b.[129]
Y-Chromosomal DNA

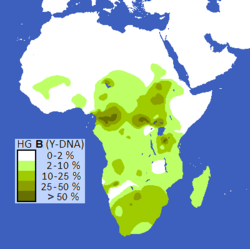
Various Y chromosome studies show that the San carry some of the most divergent (oldest) human Y-chromosome haplogroups. These haplogroups are specific sub-groups of haplogroups A and B, the two earliest branches on the human Y-chromosome tree.[134][135][136]
Mitochondrial DNA
In 200,000 BP, Africans (e.g., Khoisan of Southern Africa) bearing haplogroup L0 diverged from other Africans bearing haplogroup L1′6, which tend to be northward of Southern Africa.[83] Between 130,000 BP and 75,000 BP, behavioral modernity emerged among Southern Africans and long-term interactions between the regions of Southern Africa and Eastern Africa became established.[83]
Mitochondrial DNA studies also provide evidence that the San carry high frequencies of the earliest haplogroup branches in the human mitochondrial DNA tree. This DNA is inherited only from one's mother. The most divergent (oldest) mitochondrial haplogroup, L0d, has been identified at its highest frequencies in the southern African San groups.[134][137][138][139]
Autosomal DNA
Henn et al. (2011) found that the ǂKhomani San, as well as the Sandawe and Hadza peoples of Tanzania, were the most genetically diverse of any living humans studied. This high degree of genetic diversity hints at the origin of anatomically modern humans.[140][141]
Medical DNA
Among the ancient DNA from three hunter-gatherers sharing genetic similarity with San people and four Iron Age agriculturalists, their SNPs indicated that they bore variants for resistance against sleeping sickness and Plasmodium vivax.[142] In particular, two out of the four Iron Age agriculturalists bore variants for resistance against sleeping sickness and three out of the four Iron Age agriculturalists bore Duffy negative variants for resistance against malaria.[142] In contrast to the Iron Age agriculturalists, from among the San-related hunter-gatherers, a six-year-old boy may have died from schistosomiasis.[142] In Botswana, a man, who dates to 1400 BP, may have also carried the Duffy negative variant for resistance against malaria.[142]
The genomes of Africans commonly found to undergo adaptation are regulatory DNA, and many cases of adaptation found among Africans relate to diet, physiology, and evolutionary pressures from pathogens.[85] Throughout Sub-Saharan Africa, genetic adaptation (e.g., rs334 mutation, Duffy blood group, increased rates of G6PD deficiency, sickle cell disease) to malaria has been found among Sub-Saharan Africans, which may have initially developed in 7300 BP.[85] Sub-Saharan Africans have more than 90% of the Duffy-null genotype.[101] In the Kalahari Desert region of Africa, various possible genetic adaptations (e.g., adiponectin, body mass index, metabolism) have been found among the ǂKhomani people.[85] Sub-Saharan Africans have more than 90% of the Duffy-null genotype.[101] In South Africa , genetic adaptation (e.g., rs28647531 on chromosome 4q22) and strong susceptibility to tuberculosis has been found among Coloureds.[85]
Recent African origin of modern humans
Between 500,000 BP and 300,000 BP, anatomically modern humans may have emerged in Africa.[143] As Africans (e.g., Y-Chromosomal Adam, Mitochondrial Eve) have migrated from their places of origin in Africa to other locations in Africa, and as the time of divergence for East African, Central African, and West African lineages are similar to the time of divergence for the Southern African lineage, there is insufficient evidence to identify a specific region for the origin of humans in Africa.[73] In 100,000 BP, anatomically modern humans migrated from Africa into Eurasia.[144] Subsequently, tens of thousands of years after, the ancestors of all present-day Eurasians migrated from Africa into Eurasia and eventually became admixed with Denisovans and Neanderthals.[144]
Archaeological and fossil evidence provide support for the African origin of homo sapiens and behavioral modernity.[145] Models reflecting a pan-African origin (multiple locations of origin within Africa) and evolution of modern humans have been developed.[145] As the idea of "modern" has become increasingly problematized, research has "begun to disentangle what is meant by "modern" genetic ancestry, skeletal morphology, and behavior, recognizing these are unlikely to form a single package."[145]
In comparison to the non-African genome, the African genome features a ~25% greater number of polymorphisms,[85] or 3 to 5 times as many,[101] and genetic variants that are rare outside of Africa are found to occur at an abundant rate within Africa.[85] Most of the genetic diversity found among non-Africans is found to be, at large, a subset of genetic diversity found among Africans.[85] The genomes of Africans commonly found to undergo adaptation are regulatory DNA, and many cases of adaptation found among Africans relate to diet, physiology, and evolutionary pressures from pathogens.[85] Throughout Sub-Saharan Africa, genetic adaptation (e.g., rs334 mutation, Duffy blood group, increased rates of G6PD deficiency, sickle cell disease) to malaria has been found among Sub-Saharan Africans, which may have initially developed in 7300 BP.[85] Throughout Africa, various genetic adaptations (e.g., apolipoprotein L1 (APOL1): G1 and G2 haplotype resistance to trypanosomiasis and increased risk of kidney disease; human leukocyte antigen (HLA) genes; major histocompatibility complex (MHC)) to HIV-1, smallpox, trypanosomiasis (African sleeping sickness), and tuberculosis has been found among Africans.[85] Biomedical tests for specific genetic variants (e.g., rs1799853 in the CYP2C9 gene), which have been approved by the U.S. Food and Drug Administration and are intended to indicate correct prescription of warfarin, has been found to be increasingly irrelevant to Africans as the variants are rare in Africa.[85] As frequency rate factors into considering and deciding variant pathogenicity and generalizable polygenic scores, modern clinical classifications of genetic variant pathogenicity are found to be inadequate due to a lack of genetic diversity in biomedical studies.[85] Fan et al (2023) recently found ~5.3 million unique genetic variants in 180 African hunter-gatherer populations, and among existing classifications for variants determined to likely be “pathogenic”, ~29% (44/154) of these “pathogenic” classified variants were found to occur frequently among the African hunter-gatherers.[85]
See also
Notes
- ↑ Although the estimated date of this event varies across studies, there is general consensus that the split occurred over 100 thousand years ago (kya) (8,23).
References
- ↑ Osborne, Anne H. (October 2008). "A humid corridor across the Sahara for the migration of early modern humans out of Africa 120,000 years ago". Proceedings of the National Academy of Sciences of the United States of America 105 (43): 16444–16447. doi:10.1073/pnas.0804472105. PMID 18936490. Bibcode: 2008PNAS..10516444O.
- ↑ Drake, Nick; Breeze, Paul (2016). "Climate Change and Modern Human Occupation of the Sahara from MIS 6-2". Africa from MIS 6-2. Vertebrate Paleobiology and Paleoanthropology. Africa from MIS 6-2. pp. 103–122. doi:10.1007/978-94-017-7520-5_6. ISBN 978-94-017-7519-9. https://link.springer.com/chapter/10.1007/978-94-017-7520-5_6.
- ↑ El-Shenawy, Mohammed I. (2018). "Speleothem evidence for the greening of the Sahara and its implications for the early human dispersal out of sub-Saharan Africa". Quaternary Science Reviews 188: 67–76. doi:10.1016/j.quascirev.2018.03.016. Bibcode: 2018QSRv..188...67E. https://www.sciencedirect.com/science/article/abs/pii/S0277379117307436.
- ↑ Scheele, Judith (Aug 2016). Crossroads Regions: The Sahara. Oxford Handbooks Online. doi:10.1093/oxfordhb/9780199935369.013.18. ISBN 978-0-19-993536-9. https://www.oxfordhandbooks.com/view/10.1093/oxfordhb/9780199935369.001.0001/oxfordhb-9780199935369-e-18.
- ↑ Wippel, Steffen (2020). "The Sahara as a Bridge, Not a Barrier: An Essay and Book Review on Recent Transregional Perspectives". Neue Politische Literatur 65 (3): 449–472. doi:10.1007/s42520-020-00318-y.
- ↑ 6.0 6.1 "The deep population history in Africa". Human Molecular Genetics 30 (R1): R2–R10. April 2021. doi:10.1093/hmg/ddab005. PMID 33438014.
- ↑ 7.0 7.1 "African evolutionary history inferred from whole genome sequence data of 44 indigenous African populations". Genome Biology 20 (1): 82. April 2019. doi:10.1186/s13059-019-1679-2. PMID 31023338.
- ↑ "A great African gene migration" (in en-AU). 29 October 2020. https://cosmosmagazine.com/science/a-great-migration-of-genes-in-africa/.
- ↑ 9.0 9.1 "African genetic diversity provides novel insights into evolutionary history and local adaptations". 1 August 2018. https://academic.oup.com/hmg/article/27/R2/R209/4993963.
- ↑ Baker, Jennifer L.; Rotimi, Charles N.; Shriner, Daniel (2017-05-08). "Human ancestry correlates with language and reveals that race is not an objective genomic classifier" (in en). Scientific Reports 7 (1): 1572. doi:10.1038/s41598-017-01837-7. ISSN 2045-2322. PMID 28484253. Bibcode: 2017NatSR...7.1572B.
- ↑ Kusuma, Pradiptajati; Brucato, Nicolas; Cox, Murray P.; Pierron, Denis; Razafindrazaka, Harilanto; Adelaar, Alexander; Sudoyo, Herawati; Letellier, Thierry et al. (2016-05-18). "Contrasting Linguistic and Genetic Origins of the Asian Source Populations of Malagasy". Scientific Reports 6: 26066. doi:10.1038/srep26066. ISSN 2045-2322. PMID 27188237. Bibcode: 2016NatSR...626066K.
- ↑ "A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes". American Journal of Human Genetics 70 (5): 1197–1214. May 2002. doi:10.1086/340257. PMID 11910562.
- ↑ "Population relationships based on 170 ancestry SNPs from the combined Kidd and Seldin panels". Scientific Reports 9 (1): 18874. December 2019. doi:10.1038/s41598-019-55175-x. PMID 31827153. Bibcode: 2019NatSR...918874P.
- ↑ 14.0 14.1 14.2 "Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals". Cell 180 (4): 677–687.e16. February 2020. doi:10.1016/j.cell.2020.01.012. PMID 32004458.
- ↑ 15.0 15.1 15.2 van de Loosdrecht, Marieke; Bouzouggar, Abdeljalil; Humphrey, Louise; Posth, Cosimo; Barton, Nick; Aximu-Petri, Ayinuer; Nickel, Birgit; Nagel, Sarah et al. (2018-05-04). "Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations" (in en). Science 360 (6388): 548–552. doi:10.1126/science.aar8380. ISSN 0036-8075. PMID 29545507. Bibcode: 2018Sci...360..548V.
- ↑ "A quantitative comparison of the similarity between genes and geography in worldwide human populations". PLOS Genetics 8 (8): e1002886. August 2012. doi:10.1371/journal.pgen.1002886. PMID 22927824.
- ↑ "The demographic response to Holocene climate change in the Sahara" (in en). Quaternary Science Reviews 101: 28–35. 2014-10-01. doi:10.1016/j.quascirev.2014.07.003. ISSN 0277-3791. Bibcode: 2014QSRv..101...28M.
- ↑ Archaeology, language, and the African past. AltaMira Press. 2006. ISBN 9780759104655.
- ↑ "Can we visit the graves of the first Niger-Congo speakers?". 2nd International Congress: Towards Proto-Niger-Congo: Comparison and Reconstruction. Paris. September 2016. https://llacan.cnrs.fr/nigercongo2/discussions/Can_we_visit_the_graves_of_the_first_Niger.pdf.
- ↑ (in en) The Oxford Handbook of African Languages. Oxford University Press. 2020-03-13. ISBN 978-0-19-960989-5. https://books.google.com/books?id=wcjXDwAAQBAJ&pg=PA380.
- ↑ "Genetic Affinities among Southern Africa Hunter-Gatherers and the Impact of Admixing Farmer and Herder Populations". Molecular Biology and Evolution 36 (9): 1849–1861. September 2019. doi:10.1093/molbev/msz089. PMID 31288264.
- ↑ "Human Dispersal Out of Africa: A Lasting Debate". Evolutionary Bioinformatics Online 11 (Suppl 2): 57–68. 2016-04-21. doi:10.4137/EBO.S33489. PMID 27127403.
- ↑ "Did Our Species Evolve in Subdivided Populations across Africa, and Why Does It Matter?" (in English). Trends in Ecology & Evolution 33 (8): 582–594. August 2018. doi:10.1016/j.tree.2018.05.005. PMID 30007846.
- ↑ "One Species, Many Origins" (in en). https://www.shh.mpg.de/1474609/pan-african-origins.
- ↑ Vallini, Leonardo (7 April 2022). "Genetics and Material Culture Support Repeated Expansions into Paleolithic Eurasia from a Population Hub Out of Africa". https://academic.oup.com/gbe/article/14/4/evac045/6563828.
- ↑ "A Southeast Asian origin for present-day non-African human Y chromosomes". Human Genetics 140 (2): 299–307. February 2021. doi:10.1007/s00439-020-02204-9. PMID 32666166.
- ↑ "Recovering signals of ghost archaic introgression in African populations". Science Advances 6 (7): eaax5097. February 2020. doi:10.1126/sciadv.aax5097. PMID 32095519. Bibcode: 2020SciA....6.5097D. "Non-African populations (Han Chinese in Beijing and Utah residents with northern and western European ancestry) also show analogous patterns in the CSFS, suggesting that a component of archaic ancestry was shared before the split of African and non-African populations...One interpretation of the recent time of introgression that we document is that archaic forms persisted in Africa until fairly recently. Alternately, the archaic population could have introgressed earlier into a modern human population, which then subsequently interbred with the ancestors of the populations that we have analyzed here. The models that we have explored here are not mutually exclusive, and it is plausible that the history of African populations includes genetic contributions from multiple divergent populations, as evidenced by the large effective population size associated with the introgressing archaic population...Given the uncertainty in our estimates of the time of introgression, we wondered whether jointly analyzing the CSFS from both the CEU (Utah residents with Northern and Western European ancestry) and YRI genomes could provide additional resolution. Under model C, we simulated introgression before and after the split between African and non-African populations and observed qualitative differences between the two models in the high-frequency–derived allele bins of the CSFS in African and non-African populations (fig. S40). Using ABC to jointly fit the high-frequency–derived allele bins of the CSFS in CEU and YRI (defined as greater than 50% frequency), we find that the lower limit on the 95% credible interval of the introgression time is older than the simulated split between CEU and YRI (2800 versus 2155 generations B.P.), indicating that at least part of the archaic lineages seen in the YRI are also shared with the CEU..."
- ↑ [1] Supplementary Materials for Recovering signals of ghost archaic introgression in African populations", section "S8.2" "We simulated data using the same priors in Section S5.2, but computed the spectrum for both YRI [West African Yoruba] and CEU [a population of European origin] . We found that the best fitting parameters were an archaic split time of 27,000 generations ago (95% HPD: 26,000-28,000), admixture fraction of 0.09 (95% HPD: 0.04-0.17), admixture time of 3,000 generations ago (95% HPD: 2,800-3,400), and an effective population size of 19,700 individuals (95% HPD: 19,300-20,200). We find that the lower bound of the admixture time is further back than the simulated split between CEU and YRI (2155 generations ago), providing some evidence in favor of a pre-Out-of-Africa event. This model suggests that many populations outside of Africa should also contain haplotypes from this introgression event, though detection is difficult because many methods use unadmixed outgroups to detect introgressed haplotypes [Browning et al., 2018, Skov et al., 2018, Durvasula and Sankararaman, 2019] (5, 53, 22). It is also possible that some of these haplotypes were lost during the Out-of-Africa bottleneck."
- ↑ "Recovering signals of ghost archaic introgression in African populations". Science Advances 6 (7): eaax5097. February 2020. doi:10.1126/sciadv.aax5097. PMID 32095519. Bibcode: 2020SciA....6.5097D.
- ↑ "Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals". Cell 180 (4): 677–687.e16. February 2020. doi:10.1016/j.cell.2020.01.012. PMID 32004458.
- ↑ 31.0 31.1 Vicente, Mário; Schlebusch, Carina M (2020-06-01). "African population history: an ancient DNA perspective" (in en). Current Opinion in Genetics & Development. Genetics of Human Origin 62: 8–15. doi:10.1016/j.gde.2020.05.008. ISSN 0959-437X. PMID 32563853.
- ↑ "Admixture into and within sub-Saharan Africa". eLife 5. June 2016. doi:10.7554/eLife.15266. PMID 27324836.
- ↑ Serra-Vidal, Gerard; Lucas-Sanchez, Marcel; Fadhlaoui-Zid, Karima; Bekada, Asmahan; Zalloua, Pierre; Comas, David (2019-11-18). "Heterogeneity in Palaeolithic Population Continuity and Neolithic Expansion in North Africa" (in en). Current Biology 29 (22): 3953–3959.e4. doi:10.1016/j.cub.2019.09.050. ISSN 0960-9822. PMID 31679935.
- ↑ 34.0 34.1 "Ancient west Eurasian ancestry in southern and eastern Africa". Proceedings of the National Academy of Sciences of the United States of America 111 (7): 2632–2637. February 2014. doi:10.1073/pnas.1313787111. PMID 24550290. Bibcode: 2014PNAS..111.2632P.
- ↑ 35.0 35.1 35.2 "African genetic diversity provides novel insights into evolutionary history and local adaptations". Human Molecular Genetics 27 (R2): R209–R218. August 2018. doi:10.1093/hmg/ddy161. PMID 29741686.
- ↑ Clark, JD; Brandt, SA (1984) (in English). From Hunters to Farmers: The Causes and Consequences of Food Production in Africa. University of California Press. pp. 180. ISBN 978-0520045743.
- ↑ "Farmers and their languages: the first expansions". Science 300 (5619): 597–603. April 2003. doi:10.1126/science.1078208. PMID 12714734. Bibcode: 2003Sci...300..597D.
- ↑ Campbell, Lyle (2021) (in English). Historical Linguistics, Fourth Edition. The MIT Press. pp. 399–400. ISBN 978-0262542180.
- ↑ Jarvie; Hall (2005) (in English). Transition to Modernity: Essays on Power, Wealth and Belief. Cambridge University Press. pp. 27. ISBN 9780521022279.
- ↑ Shirai, Noriyuki (2010). The archaeology of the first farmer-herders in Egypt: new insights into the Fayum Epipalaeolithic and Neolithic. Leiden University Press. ISBN 978-90-485-1269-0. OCLC 852516752. http://worldcat.org/oclc/852516752.
- ↑ 41.0 41.1 41.2 "Early back-to-Africa migration into the Horn of Africa". PLOS Genetics 10 (6): e1004393. June 2014. doi:10.1371/journal.pgen.1004393. PMID 24921250.
- ↑ Blench, Roger (2006) (in English). Archaeology, Language, and the African Past. AltaMira Press. pp. 150–163. ISBN 978-0759104662.
- ↑ 43.0 43.1 Ehret, Christopher (1979). "On the Antiquity of Agriculture in Ethiopia". The Journal of African History 20 (2): 161–177. doi:10.1017/S002185370001700X. https://www.jstor.org/stable/181512.
- ↑ Nöth, Winfried (2011) (in English). Origins of Semiosis: Sign Evolution in Nature and Culture. De Gruyter Mouton. pp. 293. ISBN 9781134816231.
- ↑ 45.0 45.1 Shriner, Daniel (2018). "Re-analysis of Whole Genome Sequence Data From 279 Ancient Eurasians Reveals Substantial Ancestral Heterogeneity". Frontiers in Genetics 9: 268. doi:10.3389/fgene.2018.00268. ISSN 1664-8021. PMID 30079081. "and a sub-Saharan African component in Natufians that localizes to present-day southern Ethiopia.".
- ↑ Underhill, P. A.; Passarino, G.; Lin, A. A.; Shen, P.; Mirazón Lahr, M.; Foley, R. A.; Oefner, P. J.; Cavalli-Sforza, L. L. (January 2001). "The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations". Annals of Human Genetics 65 (Pt 1): 43–62. doi:10.1046/j.1469-1809.2001.6510043.x. ISSN 0003-4800. PMID 11415522.
- ↑ Ibrahim, Muntaser E. (2021-04-26). "Genetic diversity of the Sudanese: insights on origin and implications for health". Human Molecular Genetics 30 (R1): R37–R41. doi:10.1093/hmg/ddab028. ISSN 1460-2083. PMID 33864377.
- ↑ Ehret, Christopher; Keita, S. O. Y.; Newman, Paul (2004-12-03). "The origins of Afroasiatic". Science 306 (5702): 1680; author reply 1680. doi:10.1126/science.306.5702.1680c. ISSN 1095-9203. PMID 15576591.
- ↑ Ehret, Christopher (1979). "On the Antiquity of Agriculture in Ethiopia". The Journal of African History 20 (2): 161–177. doi:10.1017/S002185370001700X. ISSN 0021-8537.
- ↑ Bultosa, G.; Taylor, J. R. N. (2004-01-01), Wrigley, Colin, ed. (in en), TEFF, Oxford: Elsevier, pp. 281–290, doi:10.1016/b0-12-765490-9/00172-5, ISBN 978-0-12-765490-4, https://www.sciencedirect.com/science/article/pii/B0127654909001725, retrieved 2022-03-29
- ↑ Schlebusch, Carina M.; Jakobsson, Mattias (2018-08-31). "Tales of Human Migration, Admixture, and Selection in Africa". Annual Review of Genomics and Human Genetics 19: 405–428. doi:10.1146/annurev-genom-083117-021759. ISSN 1545-293X. PMID 29727585.
- ↑ 52.0 52.1 52.2 52.3 52.4 52.5 52.6 52.7 Schuenemann, Verena J.; Peltzer, Alexander; Welte, Beatrix; van Pelt, W. Paul; Molak, Martyna; Wang, Chuan-Chao; Furtwängler, Anja; Urban, Christian et al. (2017-05-30). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications 8 (1): 15694. doi:10.1038/ncomms15694. ISSN 2041-1723. PMID 28556824. Bibcode: 2017NatCo...815694S.
- ↑ Molinaro, Ludovica; Montinaro, Francesco; Yelmen, Burak; Marnetto, Davide; Behar, Doron M.; Kivisild, Toomas; Pagani, Luca (2019-12-11). "West Asian sources of the Eurasian component in Ethiopians: a reassessment" (in en). Scientific Reports 9 (1): 18811. doi:10.1038/s41598-019-55344-y. ISSN 2045-2322. PMID 31827175. Bibcode: 2019NatSR...918811M.
- ↑ Gandini, Francesca; Achilli, Alessandro; Pala, Maria; Bodner, Martin; Brandini, Stefania; Huber, Gabriela; Egyed, Balazs; Ferretti, Luca et al. (2016-05-05). "Mapping human dispersals into the Horn of Africa from Arabian Ice Age refugia using mitogenomes" (in en). Scientific Reports 6 (1): 25472. doi:10.1038/srep25472. ISSN 2045-2322. PMID 27146119. Bibcode: 2016NatSR...625472G.
- ↑ "Erratum for the Report "Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa" (previously titled "Ancient Ethiopian genome reveals extensive Eurasian admixture throughout the African continent") by M. Gallego Llorente, E. R. Jones, A. Eriksson, V. Siska, K. W. Arthur, J. W. Arthur, M. C. Curtis, J. T. Stock, M. Coltorti, P. Pieruccini, S. Stretton, F. Brock, T. Higham, Y. Park, M. Hofreiter, D. G. Bradley, J. Bhak, R. Pinhasi, A. Manica". Science 351 (6275): aaf3945. 2016-02-19. doi:10.1126/science.aaf3945. ISSN 1095-9203. PMID 26912899.
- ↑ López, Saioa; Tarekegn, Ayele; Band, Gavin; van Dorp, Lucy; Bird, Nancy; Morris, Sam; Oljira, Tamiru; Mekonnen, Ephrem et al. (2021-06-11). "Evidence of the interplay of genetics and culture in Ethiopia" (in en). Nature Communications 12 (1): 3581. doi:10.1038/s41467-021-23712-w. ISSN 2041-1723. PMID 34117245. Bibcode: 2021NatCo..12.3581L.
- ↑ "The culture history of Madagascar" (in en). Journal of World Prehistory 7 (4): 417–466. 1993-12-01. doi:10.1007/BF00997802.
- ↑ "A chronology for late prehistoric Madagascar". Journal of Human Evolution 47 (1–2): 25–63. 2004-07-01. doi:10.1016/j.jhevol.2004.05.005. PMID 15288523.
- ↑ "Genome-wide evidence of Austronesian-Bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar". Proceedings of the National Academy of Sciences of the United States of America 111 (3): 936–941. January 2014. doi:10.1073/pnas.1321860111. PMID 24395773. Bibcode: 2014PNAS..111..936P.
- ↑ 60.0 60.1 "Contrasting Linguistic and Genetic Origins of the Asian Source Populations of Malagasy". Scientific Reports 6 (1): 26066. May 2016. doi:10.1038/srep26066. PMID 27188237. Bibcode: 2016NatSR...626066K.
- ↑ 61.0 61.1 "Genetic evidence and historical theories of the Asian and African origins of the present Malagasy population". Human Molecular Genetics 30 (R1): R72–R78. April 2021. doi:10.1093/hmg/ddab018. PMID 33481023.
- ↑ Nicolas, Brucato (4 February 2019). "Evidence of Austronesian Genetic Lineages in East Africa and South Arabia: Complex Dispersal from Madagascar and Southeast Asia". https://academic.oup.com/gbe/article/11/3/748/5306180.
- ↑ "The genetics of East African populations: a Nilo-Saharan component in the African genetic landscape". Scientific Reports 5: 9996. May 2015. doi:10.1038/srep09996. PMID 26017457. Bibcode: 2015NatSR...5E9996D.
- ↑ "Genetic Heterogeneity between Berbers and Arabs" (in en), eLS (John Wiley & Sons, Ltd): pp. 1–7, 2017, doi:10.1002/9780470015902.a0027485, ISBN 978-0-470-01590-2
- ↑ González-Fortes, G.; Tassi, F.; Trucchi, E.; Henneberger, K.; Paijmans, J. L. A.; Díez-del-Molino, D.; Schroeder, H.; Susca, R. R. et al. (2019-01-30). "A western route of prehistoric human migration from Africa into the Iberian Peninsula". Proceedings of the Royal Society B: Biological Sciences 286 (1895): 20182288. doi:10.1098/rspb.2018.2288. ISSN 0962-8452. PMID 30963949.
- ↑ Moorjani, Priya; Patterson, Nick; Hirschhorn, Joel N.; Keinan, Alon; Hao, Li; Atzmon, Gil; Burns, Edward; Ostrer, Harry et al. (April 2011). "The history of African gene flow into Southern Europeans, Levantines, and Jews". PLOS Genetics 7 (4): e1001373. doi:10.1371/journal.pgen.1001373. ISSN 1553-7404. PMID 21533020.
- ↑ Botigué, Laura R.; Henn, Brenna M.; Gravel, Simon; Maples, Brian K.; Gignoux, Christopher R.; Corona, Erik; Atzmon, Gil; Burns, Edward et al. (2013-07-16). "Gene flow from North Africa contributes to differential human genetic diversity in southern Europe". Proceedings of the National Academy of Sciences of the United States of America 110 (29): 11791–11796. doi:10.1073/pnas.1306223110. ISSN 0027-8424. PMID 23733930. Bibcode: 2013PNAS..11011791B.
- ↑ Auton, Adam; Bryc, Katarzyna; Boyko, Adam R.; Lohmueller, Kirk E.; Novembre, John; Reynolds, Andy; Indap, Amit; Wright, Mark H. et al. (May 2009). "Global distribution of genomic diversity underscores rich complex history of continental human populations". Genome Research 19 (5): 795–803. doi:10.1101/gr.088898.108. ISSN 1088-9051. PMID 19218534.
- ↑ Reich, David; Price, Alkes L.; Patterson, Nick (May 2008). "Principal component analysis of genetic data" (in en). Nature Genetics 40 (5): 491–492. doi:10.1038/ng0508-491. ISSN 1546-1718. PMID 18443580. https://www.nature.com/articles/ng0508-491.
- ↑ Černý, Viktor; Fortes-Lima, Cesar; Tříska, Petr (2021-04-26). "Demographic history and admixture dynamics in African Sahelian populations". Human Molecular Genetics 30 (R1): R29–R36. doi:10.1093/hmg/ddaa239. ISSN 1460-2083. PMID 33105478.
- ↑ Haber, Marc; Mezzavilla, Massimo; Bergström, Anders; Prado-Martinez, Javier; Hallast, Pille; Saif-Ali, Riyadh; Al-Habori, Molham; Dedoussis, George et al. (December 2016). "Chad Genetic Diversity Reveals an African History Marked by Multiple Holocene Eurasian Migrations". The American Journal of Human Genetics 99 (6): 1316–1324. doi:10.1016/j.ajhg.2016.10.012. PMID 27889059. PMC 5142112. https://www.cell.com/ajhg/pdf/S0002-9297(16)30448-7.pdf.
- ↑ Campbell, Michael C; Hirbo, Jibril B; Townsend, Jeffrey P; Tishkoff, Sarah A (2014-12-01). "The peopling of the African continent and the diaspora into the new world". Current Opinion in Genetics & Development 29: 120–132. doi:10.1016/j.gde.2014.09.003. ISSN 0959-437X. PMID 25461616.
- ↑ 73.0 73.1 73.2 73.3 73.4 73.5 73.6 73.7 "Origins of modern human ancestry". Nature 590 (7845): 229–237. February 2021. doi:10.1038/s41586-021-03244-5. PMID 33568824. Bibcode: 2021Natur.590..229B.
- ↑ 74.0 74.1 74.2 74.3 "Insights from ancient DNA analysis of Egyptian human mummies: clues to disease and kinship". Human Molecular Genetics 30 (R1): R24–R28. April 2021. doi:10.1093/hmg/ddaa223. PMID 33059357.
- ↑ Loreille, Odile; Ratnayake, Shashikala; Bazinet, Adam L.; Stockwell, Timothy B.; Sommer, Daniel D.; Rohland, Nadin; Mallick, Swapan; Johnson, Philip L. F. et al. (March 2018). "Biological Sexing of a 4000-Year-Old Egyptian Mummy Head to Assess the Potential of Nuclear DNA Recovery from the Most Damaged and Limited Forensic Specimens". Genes 9 (3): 135. doi:10.3390/genes9030135. PMID 29494531.
- ↑ "Revisiting the harem conspiracy and death of Ramesses III: anthropological, forensic, radiological, and genetic study". BMJ 345: e8268. December 2012. doi:10.1136/bmj.e8268. PMID 23247979.
- ↑ "Ancient Egyptian Genomes from northern Egypt: Further discussion". Nature Communications. https://www.researchgate.net/publication/327065612.
- ↑ "Shocking truth behind Takabuti's death revealed" (in en). https://www.manchester.ac.uk/discover/news/shocking-truth-behind-takabutis-death-revealed/.
- ↑ 79.0 79.1 Oras, Ester; Anderson, Jaanika; Tõrv, Mari; Vahur, Signe; Rammo, Riina; Remmer, Sünne; Mölder, Maarja; Malve, Martin et al. (2020-01-16). "Multidisciplinary investigation of two Egyptian child mummies curated at the University of Tartu Art Museum, Estonia (Late/Graeco-Roman Periods)" (in en). PLOS ONE 15 (1): e0227446. doi:10.1371/journal.pone.0227446. ISSN 1932-6203. PMID 31945091. Bibcode: 2020PLoSO..1527446O.
- ↑ "Ancestral mitochondrial N lineage from the Neolithic 'green' Sahara". Scientific Reports 9 (1): 3530. March 2019. doi:10.1038/s41598-019-39802-1. PMID 30837540. Bibcode: 2019NatSR...9.3530V.
- ↑ 81.0 81.1 "Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations". Science 360 (6388): 548–552. May 2018. doi:10.1126/science.aar8380. PMID 29545507. Bibcode: 2018Sci...360..548V.
- ↑ 82.0 82.1 Jeong, Choongwon (2020). "Current Trends in Ancient DNA Study: Beyond Human Migration in and Around Europe". The Handbook of Mummy Studies. Springer. pp. 1–16. doi:10.1007/978-981-15-1614-6_10-1. ISBN 978-981-15-1614-6. OCLC 1182512815. https://link.springer.com/referenceworkentry/10.1007%2F978-981-15-1614-6_10-1.
- ↑ 83.00 83.01 83.02 83.03 83.04 83.05 83.06 83.07 83.08 83.09 83.10 83.11 83.12 83.13 83.14 83.15 83.16 83.17 83.18 83.19 Sá, Luísa (16 August 2022). "Phylogeography of Sub-Saharan Mitochondrial Lineages Outside Africa Highlights the Roles of the Holocene Climate Changes and the Atlantic Slave Trade". International Journal of Molecular Sciences 23 (16): 9219. doi:10.3390/ijms23169219. ISSN 1661-6596. OCLC 9627558751. PMID 36012483.
- ↑ "Mitochondrial HVRI and whole mitogenome sequence variations portray similar scenarios on the genetic structure and ancestry of northeast Africans". Meta Gene. http://81.95.108.158/return-files/Mitochondrial%20HVRI.pdf. Retrieved 2021-05-12.
- ↑ 85.00 85.01 85.02 85.03 85.04 85.05 85.06 85.07 85.08 85.09 85.10 85.11 85.12 85.13 85.14 85.15 85.16 85.17 85.18 85.19 85.20 85.21 85.22 85.23 85.24 Pfennig, Aaron (March 29, 2023). "Evolutionary Genetics and Admixture in African Populations". Genome Biology and Evolution 15 (4): evad054. doi:10.1093/gbe/evad054. OCLC 9817135458. PMID 36987563. PMC 10118306. https://academic.oup.com/gbe/article/15/4/evad054/7092825.
- ↑ 86.0 86.1 86.2 86.3 "Sahelian pastoralism from the perspective of variants associated with lactase persistence". American Journal of Physical Anthropology 173 (3): 423–436. November 2020. doi:10.1002/ajpa.24116. PMID 32812238. https://hal.archives-ouvertes.fr/hal-02919786/file/ajpa_ms_final.pdf.
- ↑ Scerri, Eleanor (26 October 2017). "The Stone Age Archaeology of West Africa". Oxford Research Encyclopedia of African History. Oxford University Press. p. 1. doi:10.1093/acrefore/9780190277734.013.137. ISBN 9780190277734. OCLC 1013546425. https://ora.ox.ac.uk/objects/uuid:1442d1c1-7f76-454d-97db-8fd52a4c6c1e/files/m02a1cd8d0c2ef8eb3ffee57a58c4b335.
- ↑ Batini, Chiara (September 2011). "Supplementary Data: Signatures of the Preagricultural Peopling Processes in Sub-Saharan Africa as Revealed by the Phylogeography of Early Y Chromosome Lineages". Molecular Biology and Evolution 28 (9): 2603–2613. doi:10.1093/molbev/msr089. ISSN 0737-4038. OCLC 748733133. PMID 21478374. https://academic.oup.com/mbe/article/28/9/2603/1011779#supplementary-data.
- ↑ Haber, Marc (August 2019). "A Rare Deep-Rooting D0 African Y-Chromosomal Haplogroup and Its Implications for the Expansion of Modern Humans Out of Africa". Genetics 212 (4): 1421–1428. doi:10.1534/genetics.119.302368. ISSN 0016-6731. OCLC 8291848146. PMID 31196864.
- ↑ 90.0 90.1 90.2 90.3 90.4 90.5 90.6 90.7 Shriner, Daniel; Rotimi, Charles N. (April 2018). "Whole-Genome-Sequence-Based Haplotypes Reveal Single Origin of the Sickle Allele during the Holocene Wet Phase". American Journal of Human Genetics 102 (4): 547–556. doi:10.1016/j.ajhg.2018.02.003. ISSN 0002-9297. OCLC 7353789016. PMID 29526279.
- ↑ Trombetta, Beniamino (June 2015). "Phylogeographic Refinement and Large Scale Genotyping of Human Y Chromosome Haplogroup E Provide New Insights into the Dispersal of Early Pastoralists in the African Continent". Genome Biology and Evolution 7 (7): 1940–1950. doi:10.1093/gbe/evv118. ISSN 0016-6731. OCLC 8291848146. PMID 26108492.
- ↑ 92.0 92.1 Miller, Eleanor F. (August 2018). "Global demographic history of human populations inferred from whole mitochondrial genomes". Royal Society Open Science 5 (8): 180543. doi:10.1098/rsos.180543. OCLC 8582185081. PMID 30225046. Bibcode: 2018RSOS....580543M.
- ↑ Frigi, Sabeh (August 2010). "Ancient local evolution of African mtDNA haplogroups in Tunisian Berber populations". Human Biology 82 (4): 367–384. doi:10.3378/027.082.0402. ISSN 0018-7143. OCLC 4668535228. PMID 21082907. https://www.researchgate.net/publication/47810347.
- ↑ Podgorná, Eliška (November 2013). "The genetic impact of the lake chad basin population in North Africa as documented by mitochondrial diversity and internal variation of the L3e5 haplogroup". Annals of Human Genetics 77 (6): 513–523. doi:10.1111/ahg.12040. ISSN 0003-4800. OCLC 6998015647. PMID 25069842. https://onlinelibrary.wiley.com/doi/abs/10.1111/ahg.12040.
- ↑ Busby, George BJ (June 21, 2016). "Admixture into and within sub-Saharan Africa". eLife 5: e15266. doi:10.7554/eLife.15266. PMID 27324836.
- ↑ Gurdasani, Deepti (December 3, 2014). "The African Genome Variation Project shapes medical genetics in Africa". Nature 517 (7534): 327–332. doi:10.1038/nature13997. ISSN 0028-0836. OCLC 9018039409. PMID 25470054. Bibcode: 2015Natur.517..327G.
- ↑ Choudhury, Ananyo (October 2020). "High-depth African genomes inform human migration and health". Nature 586 (7831): 741–748. doi:10.1038/s41586-020-2859-7. ISSN 0028-0836. OCLC 8691160009. PMID 33116287. Bibcode: 2020Natur.586..741C.
- ↑ Bird, Nancy (March 29, 2023). "Dense sampling of ethnic groups within African countries reveals fine-scale genetic structure and extensive historical admixture". Science Advances 9 (13): eabq2616. doi:10.1126/sciadv.abq2616. ISSN 2375-2548. OCLC 9819554112. PMID 36989356. Bibcode: 2023SciA....9.2616B.
- ↑ Fan, Shaohua; Kelly, Derek E.; Beltrame, Marcia H.; Hansen, Matthew E. B.; Mallick, Swapan; Ranciaro, Alessia; Hirbo, Jibril; Thompson, Simon et al. (2019-04-26). "African evolutionary history inferred from whole genome sequence data of 44 indigenous African populations". Genome Biology 20 (1): 82. doi:10.1186/s13059-019-1679-2. ISSN 1474-760X. PMID 31023338.
- ↑ Fan, Shaohua (March 2, 2023). "Whole-genome sequencing reveals a complex African population demographic history and signatures of local adaptation". Cell 186 (5): 923–939. doi:10.1016/j.cell.2023.01.042. ISSN 0092-8674. OCLC 9791150466. PMID 36868214. PMC 10568978. https://www.cell.com/cell/pdf/S0092-8674(23)00101-0.pdf.
- ↑ 101.0 101.1 101.2 101.3 101.4 101.5 Wonkam, Ambroise; Adeyemo, Adebowale (March 8, 2023). "Leveraging our common African origins to understand human evolution and health". Cell Genomics 3 (3): 100278. doi:10.1016/j.xgen.2023.100278. PMID 36950382. PMC 10025516. https://www.cell.com/cell-genomics/pdf/S2666-979X(23)00038-1.pdf.
- ↑ Amanzougaghene, Nadia (October 14, 2016). "High Ancient Genetic Diversity of Human Lice, Pediculus humanus, from Israel Reveals New Insights into the Origin of Clade B Lice". PLOS ONE 11 (10): e0164659. doi:10.1371/journal.pone.0164659. PMID 27741281. Bibcode: 2016PLoSO..1164659A.
- ↑ 103.0 103.1 103.2 "Evolutionary history of sickle-cell mutation: implications for global genetic medicine". Human Molecular Genetics 30 (R1): R119–R128. April 2021. doi:10.1093/hmg/ddab004. PMID 33461216.
- ↑ "Recent Adaptive Acquisition by African Rainforest Hunter-Gatherers of the Late Pleistocene Sickle-Cell Mutation Suggests Past Differences in Malaria Exposure". American Journal of Human Genetics 104 (3): 553–561. March 2019. doi:10.1016/j.ajhg.2019.02.007. PMID 30827499.
- ↑ 105.0 105.1 105.2 105.3 "Sickle β-globin haplotypes among patients with sickle cell anemia in Basra, Iraq: A cross-sectional study". Iraqi Journal of Hematology 9 (1): 23. January 2020. doi:10.4103/ijh.ijh_20_19.
- ↑ 106.0 106.1 106.2 106.3 Steverding, Dietmar (December 2020). "The spreading of parasites by human migratory activities". Virulence 11 (1): 1177–1191. doi:10.1080/21505594.2020.1809963. PMID 32862777.
- ↑ Agouti, Imane (December 2008). "Molecular basis of ß-thalassemia in Morocco: possible origins of the molecular heterogeneity". Genetic Testing 12 (4): 563–568. doi:10.1089/gte.2008.0058. PMID 18976160. https://www.cs.sjsu.edu/~khuri/Rabat_2013/Agouti_2008.pdf.
- ↑ Murunga, Philip (2018). "Mitochondrial DNA D-Loop Diversity of the Helmeted Guinea Fowls in Kenya and Its Implications on HSP70 Gene Functional Polymorphism". BioMed Research International 2018: 1–12. doi:10.1155/2018/7314038. OCLC 8754386965. PMID 30539018.
- ↑ González-Fortes, G. (2019). "A western route of prehistoric human migration from Africa into the Iberian Peninsula". Proceedings of the Royal Society B: Biological Sciences 286 (1895): 20182288. doi:10.1098/rspb.2018.2288. PMID 30963949.
- ↑ "Ancient West African foragers in the context of African population history". Nature 577 (7792): 665–670. January 2020. doi:10.1038/s41586-020-1929-1. PMID 31969706. Bibcode: 2020Natur.577..665L.
- ↑ 111.00 111.01 111.02 111.03 111.04 111.05 111.06 111.07 111.08 111.09 "Ancient genomes reveal complex patterns of population movement, interaction, and replacement in sub-Saharan Africa". Science Advances 6 (24): eaaz0183. June 2020. doi:10.1126/sciadv.aaz0183. PMID 32582847. Bibcode: 2020SciA....6..183W.
- ↑ 112.00 112.01 112.02 112.03 112.04 112.05 112.06 112.07 112.08 112.09 "Ancient genomes reveal complex patterns of population movement, interaction, and replacement in sub-Saharan Africa". Science Advances 6 (24): eaaz0183. June 2020. doi:10.1126/sciadv.aaz0183. PMID 32582847. Bibcode: 2020SciA....6..183W.
- ↑ "The genetic landscape of Equatorial Guinea and the origin and migration routes of the Y chromosome haplogroup R-V88". European Journal of Human Genetics 21 (3): 324–331. March 2013. doi:10.1038/ejhg.2012.167. PMID 22892526.
- ↑ 114.0 114.1 114.2 "Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers". Proceedings of the National Academy of Sciences of the United States of America 105 (5): 1596–1601. February 2008. doi:10.1073/pnas.0711467105. PMID 18216239. Bibcode: 2008PNAS..105.1596Q.
- ↑ Sarah A. Tishkoff et al. 2007, History of Click-Speaking Populations of Africa Inferred from mtDNA and Y Chromosome Genetic Variation. Molecular Biology and Evolution 2007 24(10):2180-2195
- ↑ Lluis Quintana-Murci et al. MtDNA diversity in Central Africa: from hunter-gathering to agriculturalism. CNRS-Institut Pasteur, Paris
- ↑ "60,000 years of interactions between Central and Eastern Africa documented by major African mitochondrial haplogroup L2". Scientific Reports 5: 12526. July 2015. doi:10.1038/srep12526. PMID 26211407. Bibcode: 2015NatSR...512526S.
- ↑ "Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies". PLOS Genetics 8 (4): e1002641. 2012. doi:10.1371/journal.pgen.1002641. PMID 22570615.
- ↑ "Genetic variation and recent positive selection in worldwide human populations: evidence from nearly 1 million SNPs". PLOS ONE 4 (11): e7888. November 2009. doi:10.1371/journal.pone.0007888. PMID 19924308. Bibcode: 2009PLoSO...4.7888L.
- ↑ "The shortness of Pygmies is associated with severe under-expression of the growth hormone receptor". Molecular Genetics and Metabolism 98 (3): 310–313. November 2009. doi:10.1016/j.ymgme.2009.05.009. PMID 19541519.
- ↑ "Ancient Ethiopian genome reveals extensive Eurasian admixture throughout the African continent". Science 350 (6262): 820–822. November 2015. doi:10.1126/science.aad2879. PMID 26449472. Bibcode: 2015Sci...350..820L.
- ↑ "Supplementary Materials for Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa". Science 350 (6262): 820–822. 13 November 2015. doi:10.1126/science.aad2879. PMID 26449472. Bibcode: 2015Sci...350..820L. https://www.science.org/doi/10.1126/science.aad2879.
- ↑ "Structure and ancestry patterns of Ethiopians in genome-wide autosomal DNA". Human Molecular Genetics 30 (R1): R42–R48. April 2021. doi:10.1093/hmg/ddab019. PMID 33547782.
- ↑ Pagani, Luca; Kivisild, Toomas; Tarekegn, Ayele; Ekong, Rosemary; Plaster, Chris; Gallego Romero, Irene; Ayub, Qasim; Mehdi, S. Qasim et al. (2012-07-13). "Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool". American Journal of Human Genetics 91 (1): 83–96. doi:10.1016/j.ajhg.2012.05.015. ISSN 1537-6605. PMID 22726845.
- ↑ 125.0 125.1 125.2 125.3 125.4 "Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa". Science 365 (6448). July 2019. doi:10.1126/science.aaw6275. PMID 31147405. Bibcode: 2019Sci...365.6275P.
- ↑ 126.0 126.1 126.2 126.3 126.4 "Supplementary Materials for Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa". Science 365 (6448): eaaw6275. 5 July 2019. doi:10.1126/science.aaw6275. PMID 31147405. Bibcode: 2019Sci...365.6275P.
- ↑ 127.0 127.1 127.2 "Reconstructing Prehistoric African Population Structure". Cell 171 (1): 59–71.e21. September 2017. doi:10.1016/j.cell.2017.08.049. PMID 28938123.
- ↑ 128.0 128.1 128.2 128.3 Brielle, Esther S. (March 29, 2023). "Supplementary Data Files for Entwined African and Asian genetic roots of medieval peoples of the Swahili coast". Nature 615 (7954): 866–873. doi:10.1038/s41586-023-05754-w. ISSN 0028-0836. OCLC 9819552636. PMID 36991187. Bibcode: 2023Natur.615..866B.
- ↑ 129.0 129.1 129.2 129.3 Lipson, Mark (23 February 2022). "Extended Data Table 1 Ancient individuals analysed in this study: Ancient DNA and deep population structure in sub-Saharan African foragers". Nature 603 (7900): 290–296. doi:10.1038/s41586-022-04430-9. ISSN 0028-0836. OCLC 9437356581. PMID 35197631. Bibcode: 2022Natur.603..290L.
- ↑ "High-resolution characterization of genetic markers in the Arabian Peninsula and Near East". University of Leeds. https://etheses.whiterose.ac.uk/6301/1/Vfernandes_PhD_thesis.pdf.
- ↑ 131.0 131.1 131.2 "Bantu-speaker migration and admixture in southern Africa". Human Molecular Genetics 30 (R1): R56–R63. April 2021. doi:10.1093/hmg/ddaa274. PMID 33367711.
- ↑ 132.0 132.1 "Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago". Science 358 (6363): 652–655. November 2017. doi:10.1126/science.aao6266. PMID 28971970. Bibcode: 2017Sci...358..652S.
- ↑ 133.0 133.1 "Supplementary Materials for Southern African ancient genomes estimate modern human divergence to 350,000to 260,000years ago". Science 358 (6363): 652–655. 3 November 2017. doi:10.1126/science.aao6266. PMID 28971970. Bibcode: 2017Sci...358..652S.
- ↑ 134.0 134.1 "African Y chromosome and mtDNA divergence provides insight into the history of click languages". Current Biology 13 (6): 464–473. March 2003. doi:10.1016/S0960-9822(03)00130-1. PMID 12646128.
- ↑ "Hierarchical patterns of global human Y-chromosome diversity". Molecular Biology and Evolution 18 (7): 1189–1203. July 2001. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- ↑ "Development of a single base extension method to resolve Y chromosome haplogroups in sub-Saharan African populations". Investigative Genetics 1 (1): 6. September 2010. doi:10.1186/2041-2223-1-6. PMID 21092339.
- ↑ "mtDNA variation in the South African Kung and Khwe-and their genetic relationships to other African populations". American Journal of Human Genetics 66 (4): 1362–1383. April 2000. doi:10.1086/302848. PMID 10739760.
- ↑ "History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation". Molecular Biology and Evolution 24 (10): 2180–2195. October 2007. doi:10.1093/molbev/msm155. PMID 17656633.
- ↑ "SNaPshot minisequencing to resolve mitochondrial macro-haplogroups found in Africa". Electrophoresis 30 (21): 3657–3664. November 2009. doi:10.1002/elps.200900197. PMID 19810027.
- ↑ "Hunter-gatherer genomic diversity suggests a southern African origin for modern humans". Proceedings of the National Academy of Sciences of the United States of America (National Academy of Sciences) 108 (13): 5154–5162. March 2011. doi:10.1073/pnas.1017511108. PMID 21383195.
- ↑ Kaplan, Matt (2011). "Gene Study Challenges Human Origins in Eastern Africa". Scientific American (Nature Publishing Group). http://www.scientificamerican.com/article/gene-study-challenges-human-origin-africa/. Retrieved 22 June 2012.
- ↑ 142.0 142.1 142.2 142.3 "Disease as a Factor in the African Archaeological Record". The African Archaeological Review 37 (3): 487–490. 2020. doi:10.1007/s10437-020-09405-7. PMID 32863518.
- ↑ "African genetic diversity and adaptation inform a precision medicine agenda". Nature Reviews. Genetics (Nature Reviews) 22 (5): 284–306. May 2021. doi:10.1038/s41576-020-00306-8. PMID 33432191.
- ↑ 144.0 144.1 "The influence of evolutionary history on human health and disease". Nature Reviews. Genetics 22 (5): 269–283. May 2021. doi:10.1038/s41576-020-00305-9. PMID 33408383.
- ↑ 145.0 145.1 145.2 Prendergast, Mary E.; Sawchuk, Elizabeth A.; Sirak, Kendra A. (19 October 2022). "Genetics and the African Past". Oxford Research Encyclopedia of African History. Oxford University Press. ISBN 9780190277734. OCLC 1013546425. https://oxfordre.com/africanhistory/display/10.1093/acrefore/9780190277734.001.0001/acrefore-9780190277734-e-143.
 |
