Biology:Haplogroup E-M96
| Haplogroup E | |
|---|---|
| Possible time of origin | 65,200 years BP,[1] 69,000 years BP,[2] or 73,000 years BP[3] |
| Coalescence age | 52,300 years BP[1] |
| Possible place of origin | East Africa,[4][5][6][3] West Africa,[7] or Eurasia[8][9] |
| Ancestor | DE |
| Descendants | E-P147, E-M75 |
| Defining mutations | L339, L614, M40/SRY4064/SRY8299, M96, P29, P150, P152, P154, P155, P156, P162, P168, P169, P170, P171, P172, P173, P174, P175, P176 |
Haplogroup E-M96 is a human Y-chromosome DNA haplogroup. It is one of the two main branches of the older and ancestral haplogroup DE, the other main branch being haplogroup D. The E-M96 clade is divided into two main subclades: the more common E-P147, and the less common E-M75.
Origins
Underhill (2001) proposed that haplogroup E may have arisen in East Africa.[10] Some authors as Chandrasekar (2007), accept the earlier position of Hammer (1997) that Haplogroup E may have originated in West Asia,[11] given that:
- E is a clade of haplogroup DE, with the other major clade, haplogroup D, being exclusively distributed in Asia.
- DE is a clade within M168 with the other two major clades, C and F, considered to have already a Eurasian origin.
However, several discoveries made since the Hammer articles are thought to make an Asian origin less likely:
- Underhill and Kivisild (2007) demonstrated that C and F have a common ancestor meaning that DE has only one sibling which is non-African.[12]
- DE* is found in both Asia and Africa, meaning that not only one, but several siblings of D are found in Asia and Africa.
- Karafet (2008), in which Hammer is a co-author, significantly rearranged time estimates leading to "new interpretations on the geographical origin of ancient sub-clades".[13] Amongst other things this article proposed a much older age for haplogroup E-M96 than had been considered previously, giving it a similar age to Haplogroup D, and DE itself, meaning that there is no longer any strong reason to see it as an offshoot of DE which must have happened long after DE came into existence and had entered Asia.[13]
Kohl et al. (2009) presumed a West African origin for haplogroup E, stating: "From the 20 main haplogroups in the Y-chromosomal haplogroup tree, only 5 were detected in the analysed Amharic population in Ethiopia. Haplogroup A is near the roots of the tree and is only found among males on the African continent. The major haplogroup detected was E. Haplogroup E has its origin in West Africa. Due to immigration haplogroup A, which originally dominated in Ethiopia, has been partly replaced."[7]
In 2015, Poznik & Underhill et al. claimed haplogroup E arose outside Africa, arguing that, "This model of geographical segregation within the CT clade requires just one continental haplogroup exchange (E to Africa), rather than three (D, C, and F out of Africa). The timing of this putative return to Africa, between the emergence of haplogroup E and its differentiation within Africa by 58 kya, is consistent with proposals, based on non–Y chromosome data, of abundant gene flow between Africa and Arabia 50–80 kya."[8]
In 2015, Trobetta et al. suggested an East African origin for haplogroup E, stating: "our phylogeographic analysis, based on thousands of samples worldwide, suggests that the radiation of haplogroup E started about 58 ka, somewhere in sub-Saharan Africa, with a higher posterior probability (0.73) for an eastern African origin."[6]
Cabrera et al. (2018) hypothesizes a Eurasian center of origin and dispersal for haplogroup E based on the similar age of the clade's parent haplogroup DE and the mtDNA haplogroup L3. According to this hypothesis, after an initial Out-of-Africa migration of early anatomically modern humans around 125 kya, haplogroup DE diversified around the Himalayas and in or westward of the Tibet, after which E-carrying males are proposed to have back-migrated from the paternal haplogroup's place of origin in Eurasia around 70 kya along with females bearing the maternal haplogroup L3, which the study also hypothesizes to have originated in Eurasia, into Africa. These new Eurasian lineages were then suggested to have largely replaced the old autochthonous male (such as haplogroup B-M60) and female African lineages.[9]
Haber et al. (2019) study proposed an African origin for haplogroup E based on an analysis of the Y-chromosomal phylogenetic structure, haplogroup divergence times, and the recently discovered haplogroup D0 found in three Nigerians, an additional branch of the DE lineage diverging early from haplogroup D. The authors support an African origin for haplogroup DE, and the immigration of haplogroups C, D and FT out of Africa around 50,300–81,000 ybp. The early divergence dates found in the study for DE, E, and D0 (all dated to about 71-76 kya), which are determined to predate the migration out-of-Africa of the ancestors of Eurasians (dated to ca. 50-60 kya), are also considered by the authors to support an African origin for those haplogroups.[3]
Ancient DNA
Pre-Pottery Neolithic B remains from the Levant were found to have carried haplogroup E (1/7; ~14%).[14]
At Nyarindi Rockshelter, in Kenya, there were two individuals, dated to the Later Stone Age (3500 BP); one carried haplogroup L4b2a and another carried haplogroup E (E-M96, E-P162).[15][16]
At Kindoki, in the Democratic Republic of Congo, there were three individuals, dated to the protohistoric period (230 BP, 150 BP, 230 BP); one carried haplogroups E1b1a1a1d1a2 (E-CTS99, E-CTS99) and L1c3a1b, another carried haplogroup E (E-M96, E-PF1620), and the last carried haplogroups R1b1 (R-P25 1, R-M415) and L0a1b1a1.[15][16]
Distribution
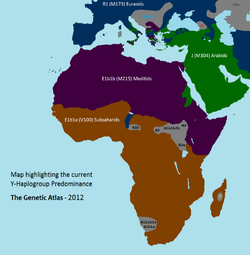
Most members of haplogroup E-M96 belong to E1, while haplogroup E2-M75 is rare. Haplogroup E1a is split into two main branches: E1a1 (E-M44) which has been mostly found in Europe, West Asia and among Ashkenazi Jews; and E1a2 (E-Z958) which has been exclusively identified in Sub-Saharan Africa.[17] Haplogroup E-M2 is the most prevalent subclade of E in Sub-Saharan Africa and is strongly associated with expansion of Bantu speakers.
E-M215 is found at high frequencies in North Africa, West Asia, East Africa and Europe. E-M215 is most common among Afro-Asiatic speakers in the Near East, North Africa and the Horn of Africa, and it has also been reported among some Nilo-Saharan and Niger–Congo speakers in North East Africa and Sudan. E-M215 is far less common in West, Central, and Southern Africa, though it has been observed among some Khoisan speakers[18] and among Niger–Congo speakers in Senegambia,[19] Guinea-Bissau,[20] Burkina Faso,[21] Ghana, Gabon,[22] the Democratic Republic of the Congo, Rwanda,[23] Namibia, and South Africa .
Subclades
E-M96*
Paragroup E-M96* refers to lineages belonging to the E clade but which cannot be classified into any known branch. E(xE1-P147, E2-M75) - that is, E which has tested negative for both P147 and M75 - has been reported in 3 males from Lebanon,[24] 2 Amharas from Ethiopia,[25] 2 males from Syria,[26] 2 males from Saudi Arabia,[27] and in a single Bantu-speaking male from South Africa .[13] E(xE1a-M33, E1b1-P2, E2-M75) was reported among several Southern African populations and in an Egyptian man;[28] E(xE1a-M33, E1b1a1-M2, E1b1b-M215, E2-M75) has also been observed amongst pygmies and Bantu from Cameroon and Gabon;[22] and also in Burkina Faso[29] and a Fulbe man from Niger.[30]
Recently it was discovered that 3 East African men previously classified only as E*-M96 could be assigned to a new branch, E-V44, which is a sister branch to E1-P147; E-P147 and E-V44 share the V3725 mutation, making E2-M75 and E-V3725 the two known primary branches of E.[31] Two Saudi private testers from Mecca and Jizan were also found to belong to this elusive and rare branch.[32] It is not known whether or not some (or all) other E*(xE1,E2) in previous studies would fall into V44 as well.
E-P147
E-P147 (also known as E1) is by far the most numerous and widely distributed branch of E-M96. It has two primary branches: E-M132 (E1a) and E-P177 (E1b). Haplogroup E1a is split into two branches: E1a1 (E-M44) which has been mostly found in Europe, West Asia and among Ashkenazi Jews; and E1a2 (E-Z958) which has been exclusively identified in Sub-Saharan Africa.[33]
Haplogroup E-P2 (E1b1) is the most frequent variant of E-M96 and the most common Y-DNA lineage in Africa with two main descendants: E-V38 (E1b1a) and E-M215 (E1b1b). Haplogroup E (xE3b,E3a) - that is, E tested negative for both M35 and M2, has been reported in 11 males from Morocco in Zalloua et al. (2008b).[34]
Haplogroup E-V38 is the ancestor of E-M2 (E1b1a1) which is the most common subclade of E in the entirety of Sub-Saharan Africa, and is strongly associated with the expansion of Bantu speakers throughout Central and Southern Africa. Another descendant of E-V38, E-M329 (E1b1a2), has been observed in an Ethiopian hunter-gatherer from 4,200 ybp, and is mostly found in males from the Horn of Africa and Arabian Peninsula.[35]
On the other hand, haplogroup E-M215 (E1b1b) is distributed in high frequencies throughout North Africa, Western Asia, East Africa and Europe. Haplogroup E-Z827 was found in Natufian samples (E-Z830+) dated to 10,000 ybp from Palestine, and is commonly found throughout West Asia, North Africa, Europe and Ethiopia. Haplogroup E-V68 is also commonly observed in North Africa and West Asia, and has been found in Iberomaurusian remains dating to 15,000 ybp from Morocco, with its prolific downstream descendant E-V32 dominating male lineages in Horn of Africa.
E-M75
E-M75 (also known as E2) is present throughout Subequatorial Africa, particularly in the African Great Lakes and Central Africa. The highest concentration of the haplogroup has been found among the Alur (66.67%),[28] Hema (38.89%),[28] Rimaibe (27.03%),[21] Mbuti (25.00%),[21] Daba (22.22%),[21] Eviya (20.83%),[22] Zulu (20.69%),[28] and Kenyan Bantus (17.24%).[23]
Haplogroup E-M75(xM41,M54) has been found in 6% (1/18) of Dama from Namibia,[28] 4% (1/26) of Ganda from Uganda,[28] 3% (1/39) of Mandinka from Gambia/Senegal,[28] and 2% (1/49) of Shona from Zimbabwe.[28]
Private commercial DNA testing at Family Tree DNA shows numerous E-M75 males originating from the Arabian Peninsula (Saudi Arabia, Bahrain, Kuwait, Yemen, and the United Arab Emirates), and among Ashkenazi Jews.[36] E-M75 has also been identified in a Lebanese male.[37]
Phylogenetics
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E-P29 | 21 | III | 3A | 13 | Eu3 | H2 | B | E* | E | E | E | E | E | E | E | E | E | E |
| E-M33 | 21 | III | 3A | 13 | Eu3 | H2 | B | E1* | E1 | E1a | E1a | E1 | E1 | E1a | E1a | E1a | E1a | E1a |
| E-M44 | 21 | III | 3A | 13 | Eu3 | H2 | B | E1a | E1a | E1a1 | E1a1 | E1a | E1a | E1a1 | E1a1 | E1a1 | E1a1 | E1a1 |
| E-M75 | 21 | III | 3A | 13 | Eu3 | H2 | B | E2a | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 | E2 |
| E-M54 | 21 | III | 3A | 13 | Eu3 | H2 | B | E2b | E2b | E2b | E2b1 | - | - | - | - | - | - | - |
| E-P2 | 25 | III | 4 | 14 | Eu3 | H2 | B | E3* | E3 | E1b | E1b1 | E3 | E3 | E1b1 | E1b1 | E1b1 | E1b1 | E1b1 |
| E-M2 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a* | E3a | E1b1 | E1b1a | E3a | E3a | E1b1a | E1b1a | E1b1a | E1b1a1 | E1b1a1 |
| E-M58 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a1 | E3a1 | E1b1a1 | E1b1a1 | E3a1 | E3a1 | E1b1a1 | E1b1a1 | E1b1a1 | E1b1a1a1a | E1b1a1a1a |
| E-M116.2 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a2 | E3a2 | E1b1a2 | E1b1a2 | E3a2 | E3a2 | E1b1a2 | E1b1a2 | E1ba12 | removed | removed |
| E-M149 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a3 | E3a3 | E1b1a3 | E1b1a3 | E3a3 | E3a3 | E1b1a3 | E1b1a3 | E1b1a3 | E1b1a1a1c | E1b1a1a1c |
| E-M154 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a4 | E3a4 | E1b1a4 | E1b1a4 | E3a4 | E3a4 | E1b1a4 | E1b1a4 | E1b1a4 | E1b1a1a1g1c | E1b1a1a1g1c |
| E-M155 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a5 | E3a5 | E1b1a5 | E1b1a5 | E3a5 | E3a5 | E1b1a5 | E1b1a5 | E1b1a5 | E1b1a1a1d | E1b1a1a1d |
| E-M10 | 8 | III | 5 | 15 | Eu2 | H2 | B | E3a6 | E3a6 | E1b1a6 | E1b1a6 | E3a6 | E3a6 | E1b1a6 | E1b1a6 | E1b1a6 | E1b1a1a1e | E1b1a1a1e |
| E-M35 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b* | E3b | E1b1b1 | E1b1b1 | E3b1 | E3b1 | E1b1b1 | E1b1b1 | E1b1b1 | removed | removed |
| E-M78 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b1* | E3b1 | E1b1b1a | E1b1b1a1 | E3b1a | E3b1a | E1b1b1a | E1b1b1a | E1b1b1a | E1b1b1a1 | E1b1b1a1 |
| E-M148 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b1a | E3b1a | E1b1b1a3a | E1b1b1a1c1 | E3b1a3a | E3b1a3a | E1b1b1a3a | E1b1b1a3a | E1b1b1a3a | E1b1b1a1c1 | E1b1b1a1c1 |
| E-M81 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2* | E3b2 | E1b1b1b | E1b1b1b1 | E3b1b | E3b1b | E1b1b1b | E1b1b1b | E1b1b1b | E1b1b1b1 | E1b1b1b1a |
| E-M107 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2a | E3b2a | E1b1b1b1 | E1b1b1b1a | E3b1b1 | E3b1b1 | E1b1b1b1 | E1b1b1b1 | E1b1b1b1 | E1b1b1b1a | E1b1b1b1a1 |
| E-M165 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b2b | E3b2b | E1b1b1b2 | E1b1b1b1b1 | E3b1b2 | E3b1b2 | E1b1b1b2a | E1b1b1b2a | E1b1b1b2a | E1b1b1b2a | E1b1b1b1a2a |
| E-M123 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b3* | E3b3 | E1b1b1c | E1b1b1c | E3b1c | E3b1c | E1b1b1c | E1b1b1c | E1b1b1c | E1b1b1c | E1b1b1b2a |
| E-M34 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b3a* | E3b3a | E1b1b1c1 | E1b1b1c1 | E3b1c1 | E3b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1c1 | E1b1b1b2a1 |
| E-M136 | 25 | III | 4 | 14 | Eu4 | H2 | B | E3b3a1 | E3b3a1 | E1b1b1c1a | E1b1b1c1a1 | E3b1c1a | E3b1c1a | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1c1a1 | E1b1b1b2a1a1 |
Research publications
The following research teams per their publications were represented in the creation of the YCC tree.
Phylogenetic trees
This phylogenetic tree of haplogroup subclades is based on the Y-Chromosome Consortium (YCC) Tree,[38] the ISOGG Y-DNA Haplogroup Tree,[39] and subsequent published research.
| E (M96) |
| ||||||||||||||||||||||||||||||
See also
Genetics
Y-DNA E subclades
Y-DNA backbone tree
References
- ↑ 1.0 1.1 "E YTree". https://www.yfull.com/tree/E/.
- ↑ "A recent bottleneck of Y chromosome diversity coincides with a global change in culture". Genome Research 25 (4): 459–466. April 2015. doi:10.1101/gr.186684.114. PMID 25770088.
- ↑ 3.0 3.1 3.2 "A Rare Deep-Rooting D0 African Y-chromosomal Haplogroup and its Implications for the Expansion of Modern Humans Out of Africa". Genetics 212 (4): 1421–1428. June 2019. doi:10.1534/genetics.119.302368. PMID 31196864.
- ↑ Semino, Ornella; Magri, Chiara; Benuzzi, Giorgia; Lin, Alice A.; Al-Zahery, Nadia; Battaglia, Vincenza; MacCioni, Liliana; Triantaphyllidis, Costas et al. (2004). "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area". The American Journal of Human Genetics 74 (5): 1023–34. doi:10.1086/386295. PMID 15069642.
- ↑ Chiaroni, J.; Underhill, P. A.; Cavalli-Sforza, L. L. (2009). "Y chromosome diversity, human expansion, drift, and cultural evolution". Proceedings of the National Academy of Sciences 106 (48): 20174–9. doi:10.1073/pnas.0910803106. PMID 19920170. Bibcode: 2009PNAS..10620174C.
- ↑ 6.0 6.1 Trombetta et al. 2015, Phylogeographic refinement and large scale genotyping of human Y chromosome haplogroup E provide new insights into the dispersal of early pastoralists in the African continent
- ↑ 7.0 7.1 Kohl, M. (November 13, 2009). "Distribution of Y-chromosomal SNP-haplogroups between males from Ethiopia". Forensic Science International: Genetics Supplement Series 2 (1): P435-436. doi:10.1016/j.fsigss.2009.08.051. ISSN 1875-1768. OCLC 4934346091. https://www.fsigeneticssup.com/article/S1875-1768(09)00099-7/fulltext.
- ↑ 8.0 8.1 Poznik, G David (2016). "Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences". Nature Genetics 48 (6): 593–599. doi:10.1038/ng.3559. PMID 27111036.
- ↑ 9.0 9.1 Cabrera, Vicente; Marrero, Patricia; Abu-Amero, Khaled; Larruga, Jose (2019). "Carriers of mitochondrial DNA macrohaplogroup L3 basal lineages migrated back to Africa from Asia around 70,000 years ago". BMC Evolutionary Biology 18 (98): 98. doi:10.1186/s12862-018-1211-4. PMID 29921229.
- ↑ Underhill, P. A.; Passarino, G.; Lin, A. A.; Shen, P.; Mirazon Lahr, M.; Foley, R. A.; Oefner, P. J.; Cavalli-Sforza, L. L. (2001). "The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations". Annals of Human Genetics 65 (Pt 1): 43–62. doi:10.1046/j.1469-1809.2001.6510043.x. PMID 11415522.
- ↑ Chandrasekar et al. (Sep–Oct 2007). "YAP insertion signature in South Asia". Annals of Human Biology 34 (5): 582–6. doi:10.1080/03014460701556262. PMID 17786594.
- ↑ Underhill, Peter A.; Kivisild, Toomas (2007). "Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations". Annual Review of Genetics 41: 539–64. doi:10.1146/annurev.genet.41.110306.130407. PMID 18076332.
- ↑ 13.0 13.1 13.2 Karafet, T. M.; Mendez, F. L.; Meilerman, M. B.; Underhill, P. A.; Zegura, S. L.; Hammer, M. F. (2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–8. doi:10.1101/gr.7172008. PMID 18385274.
- ↑ Martiniano, Rui; Sanctis, Bianca De; Hallast, Pille; Durbin, Richard (20 December 2020). "Supplementary Material: Placing ancient DNA sequences into reference phylogenies". Molecular Biology and Evolution 39 (2): 2020.12.19.423614. doi:10.1093/molbev/msac017. PMID 35084493.
- ↑ 15.0 15.1 Wang, Ke (2020). "Ancient genomes reveal complex patterns of population movement, interaction, and replacement in sub-Saharan Africa". Science Advances 6 (24): eaaz0183. doi:10.1126/sciadv.aaz0183. ISSN 2375-2548. OCLC 8616876709. PMID 32582847. Bibcode: 2020SciA....6..183W.
- ↑ 16.0 16.1 Wang, Ke (2020). "Supplementary Materials for Ancient genomes reveal complex patterns of population movement, interaction, and replacement in sub-Saharan Africa". Science Advances 6 (24): eaaz0183. doi:10.1126/sciadv.aaz0183. ISSN 2375-2548. OCLC 8616876709. PMID 32582847. PMC 7292641. Bibcode: 2020SciA....6..183W. https://advances.sciencemag.org/content/suppl/2020/06/08/6.24.eaaz0183.DC1/aaz0183_SM.pdf.
- ↑ E1a-M132 (Yfull)
- ↑ Cruciani, Fulvio; La Fratta, Roberta; Santolamazza, Piero; Sellitto, Daniele; Pascone, Roberto; Moral, Pedro; Watson, Elizabeth; Guida, Valentina et al. (2004). "Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out of Africa". The American Journal of Human Genetics 74 (5): 1014–22. doi:10.1086/386294. PMID 15042509.
- ↑ Wood, Elizabeth T; Stover, Daryn A; Ehret, Christopher; Destro-Bisol, Giovanni; Spedini, Gabriella; McLeod, Howard; Louie, Leslie; Bamshad, Mike et al. (2005). "Contrasting patterns of Y chromosome and mtDNA variation in Africa: Evidence for sex-biased demographic processes". European Journal of Human Genetics 13 (7): 867–76. doi:10.1038/sj.ejhg.5201408. PMID 15856073. (cf. Appendix A: Y Chromosome Haplotype Frequencies)
- ↑ Rosa, Alexandra; Ornelas, Carolina; Jobling, Mark A; Brehm, António; Villems, Richard (2007). "Y-chromosomal diversity in the population of Guinea-Bissau: A multiethnic perspective". BMC Evolutionary Biology 7: 124. doi:10.1186/1471-2148-7-124. PMID 17662131.
- ↑ 21.0 21.1 21.2 21.3 Cruciani, Fulvio; Santolamazza, Piero; Shen, Peidong; MacAulay, Vincent; Moral, Pedro; Olckers, Antonel; Modiano, David; Holmes, Susan et al. (2002). "A Back Migration from Asia to Sub-Saharan Africa is Supported by High-Resolution Analysis of Human Y-Chromosome Haplotypes". The American Journal of Human Genetics 70 (5): 1197–214. doi:10.1086/340257. PMID 11910562.
- ↑ 22.0 22.1 22.2 Berniell-Lee, G.; Calafell, F.; Bosch, E.; Heyer, E.; Sica, L.; Mouguiama-Daouda, P.; Van Der Veen, L.; Hombert, J.-M. et al. (2009). "Genetic and Demographic Implications of the Bantu Expansion: Insights from Human Paternal Lineages". Molecular Biology and Evolution 26 (7): 1581–9. doi:10.1093/molbev/msp069. PMID 19369595.
- ↑ 23.0 23.1 Luis, J; Rowold, D; Regueiro, M; Caeiro, B; Cinnioglu, C; Roseman, C; Underhill, P; Cavallisforza, L et al. (2004). "The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations". The American Journal of Human Genetics 74 (3): 532–44. doi:10.1086/382286. PMID 14973781.
- ↑ Zalloua, Pierre (2008). "Y-Chromosomal Diversity in Lebanon Is Structured by Recent Historical Events". Am J Hum Genet 82 (4): 873–882. doi:10.1016/j.ajhg.2008.01.020. PMID 18374297.
- ↑ Abu-Amero, Khaled K; Hellani, Ali; González, Ana M; Larruga, Jose M; Cabrera, Vicente M; Underhill, Peter A (2011). "Variation in Y chromosome, mitochondrial DNA and labels of identity on Ethiopia". UCL Discovery 10: 59. doi:10.1186/1471-2156-10-59. PMID 19772609.
- ↑ Zalloua, Pierre (2008). "Identifying Genetic Traces of Historical Expansions: Phoenician Footprints in the Mediterranean". Am J Hum Genet 83 (5): 633–642. doi:10.1016/j.ajhg.2008.10.012. PMID 18976729.
- ↑ Abu-Amero, Khaled K; Hellani, Ali; González, Ana M; Larruga, Jose M; Cabrera, Vicente M; Underhill, Peter A (2009). "Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions". BMC Genetics 10: 59. doi:10.1186/1471-2156-10-59. PMID 19772609.
- ↑ 28.0 28.1 28.2 28.3 28.4 28.5 28.6 28.7 Wood (2005). "Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes". Eur J Hum Genet 13 (7): 867–76. doi:10.1038/sj.ejhg.5201408. PMID 15856073.
- ↑ de Filippo (2011). "Y-chromosomal variation in Sub-Saharan Africa: insights into the history of Niger-Congo groups". Mol Biol Evol 28 (3): 1255–1269. doi:10.1093/molbev/msq312. PMID 21109585.
- ↑ Bučkova (2013). "Multiple and differentiated contributions to the male gene pool of pastoral and farmer populations of the African Sahel". Am J Phys Anthropol 151 (1): 10–21. doi:10.1002/ajpa.22236. PMID 23460272.
- ↑ Trombetta (2015). "Phylogeographic refinement and large scale genotyping of human Y chromosome haplogroup E provide new insights into the dispersal of early pastoralists in the African continent". Genome Biol Evol 7 (7): 1940–50. doi:10.1093/gbe/evv118. PMID 26108492.
- ↑ E-FTA45776
- ↑ E1a-M132
- ↑ Zalloua, Pierre (2008). "Identifying Genetic Traces of Historical Expansions: Phoenician Footprints in the Mediterranean". Am J Hum Genet 83 (5): 633–642. doi:10.1016/j.ajhg.2008.10.012. PMID 18976729.
- ↑ E-M329
- ↑ Family Tree DNA public haplotree, Haplogroup E-M75
- ↑ Platt, D.E., Artinian, H., Mouzaya, F. et al. Autosomal genetics and Y-chromosome haplogroup L1b-M317 reveal Mount Lebanon Maronites as a persistently non-emigrating population. Eur J Hum Genet 29, 581–592 (2021). 10.1038/s41431-020-00765-x
- ↑ Krahn, Thomas. "YCC Tree". Houston, Texas: FTDNA. http://ytree.ftdna.com/.
- ↑ International Society of Genetic Genealogy. "Y-DNA Haplogroup Tree". http://www.isogg.org/tree/ISOGG_YDNATreeTrunk.html.
Sources for conversion tables
- Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H. et al. (February 2001). "A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania". The American Journal of Human Genetics 68 (2): 432–443. doi:10.1086/318205. PMID 11170891.
- Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi et al. (1 July 2001). "Hierarchical Patterns of Global Human Y-Chromosome Diversity". Molecular Biology and Evolution 18 (7): 1189–1203. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- Jobling, Mark A.; Tyler-Smith, Chris (2000), "New uses for new haplotypes", Trends in Genetics 16 (8): 356–62, doi:10.1016/S0168-9525(00)02057-6, PMID 10904265
- Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora et al. (February 2001). "Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages". European Journal of Human Genetics 9 (2): 97–104. doi:10.1038/sj.ejhg.5200597. PMID 11313742.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William et al. (September 2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics 69 (3): 615–628. doi:10.1086/323299. PMID 11481588.
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science 290 (5494): 1155–9, doi:10.1126/science.290.5494.1155, PMID 11073453, Bibcode: 2000Sci...290.1155S
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics 65 (6): 1718–1724. doi:10.1086/302680. PMID 10577926.
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics 26 (3): 358–361. doi:10.1038/81685. PMID 11062480.
Further reading
- Arredi, B; Poloni, E; Paracchini, S; Zerjal, T; Fathallah, D; Makrelouf, M; Pascali, V; Novelletto, A et al. (2004). "A Predominantly Neolithic Origin for Y-Chromosomal DNA Variation in North Africa". The American Journal of Human Genetics 75 (2): 338–45. doi:10.1086/423147. PMID 15202071.
- Cruciani, Fulvio; Santolamazza, Piero; Shen, Peidong; MacAulay, Vincent; Moral, Pedro; Olckers, Antonel; Modiano, David; Holmes, Susan et al. (2002). "A Back Migration from Asia to Sub-Saharan Africa is Supported by High-Resolution Analysis of Human Y-Chromosome Haplotypes". The American Journal of Human Genetics 70 (5): 1197–214. doi:10.1086/340257. PMID 11910562.
- Cruciani, F; La Fratta, R.; Trombetta, B; Santolamazza, P; Sellitto, D; Colomb, EB; Dugoujon, JM; Crivellaro, F et al. (2007). "Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12". Molecular Biology and Evolution 24 (6): 1300–11. doi:10.1093/molbev/msm049. PMID 17351267. Also see Supplementary Data.
- Hammer, MF; Blackmer, F; Garrigan, D; Nachman, MW; Wilder, JA (2003). "Human population structure and its effects on sampling Y chromosome sequence variation". Genetics 164 (4): 1495–1509. doi:10.1093/genetics/164.4.1495. PMID 12930755.
- Karafet, TM; Mendez, FL; Meilerman, MB; Underhill, PA; Zegura, SL; Hammer, MF (May 2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research 18 (5): 830–8. doi:10.1101/gr.7172008. PMID 18385274. PMC 2336805. http://www.genome.org/cgi/content/abstract/gr.7172008v1.. Published online April 2, 2008. See also Supplementary Material.
- Sanchez, JJ; Hallenberg, C; Børsting, C; Hernandez, A; Morling, N (2005). "High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males". European Journal of Human Genetics 13 (7): 856–866. doi:10.1038/sj.ejhg.5201390. PMID 15756297.. Published online 9 March 2005
- Trombetta (2015). "Phylogeographic Refinement and Large Scale Genotyping of Human Y Chromosome Haplogroup E Provide New Insights into the Dispersal of Early Pastoralists in the African Continent". Genome Biology and Evolution 7 (7): 1940–1950. doi:10.1093/gbe/evv118. PMID 26108492. PMC 4524485. https://academic.oup.com/gbe/article/7/7/1940/631621.
- Wood, E. T. (2005). "Contrasting patterns of Y chromosome and mtDNA variation in Africa: evidence for sex-biased demographic processes". European Journal of Human Genetics 13 (7): 867–876. doi:10.1038/sj.ejhg.5201408. PMID 15856073.
External links
Phylogenetic tree and distribution maps of Y-DNA haplogroup E
- Y-DNA Haplogroup E and Its Subclades from ISOGG 2008
- Map of E1b1b1 distribution in Europe
- Distribution of E1b1a/E3a in Africa
- Frequency Distributions of Y-DNA Haplogroup E and its subclades - with Video Tutorial
Projects
- Haplogroup E-V38 Y-DNA Project at FTDNA
- E-M243 Y-DNA Project at FTDNA
- Haplozone::The E-M35 Phylogeny Project (former E3b Project)
- Jewish E-M243 Project at FTDNA
 |