Biology:Nucleoside-triphosphatase
| nucleoside-triphosphatase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 3.6.1.15 | ||||||||
| CAS number | 9075-51-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
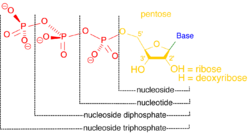
In enzymology, a nucleoside-triphosphatase (NTPase) (EC 3.6.1.15) is an enzyme that catalyzes the chemical reaction
- NTP + H2O [math]\displaystyle{ \rightleftharpoons }[/math] NDP + phosphate
Thus, the two substrates of this enzyme are NTP and H2O, whereas its two products are NDP and phosphate.
This enzyme belongs to the family of hydrolases, specifically those acting on acid anhydrides in phosphorus-containing anhydrides. The systematic name of this enzyme class is unspecific diphosphate phosphohydrolase. Other names in common use include nucleoside triphosphate phosphohydrolase, nucleoside-5-triphosphate phosphohydrolase, and nucleoside 5-triphosphatase. This enzyme participates in purine metabolism and thiamine metabolism.
Structural studies
As of late 2007, only one structure has been solved for this class of enzymes, with the PDB accession code 2I3B.
References
- "Lysosomal acid pyrophosphatase and acid phosphatase". Arch. Biochem. Biophys. 124 (1): 333–43. 1968. doi:10.1016/0003-9861(68)90335-4. PMID 5661608.
- "Properties of a soluble nucleoside triphosphatase activity in mammalian liver". Arch. Biochem. Biophys. 109 (3): 490–8. 1965. doi:10.1016/0003-9861(65)90393-0. PMID 14320490.
- "Purification of nucleoside triphosphatases from pea seedling ribosomes". Biochim. Biophys. Acta 166 (3): 707–10. 1968. doi:10.1016/0005-2787(68)90380-8. PMID 4301913.
 |

