Biology:Amyloid

Amyloids are aggregates of proteins characterised by a fibrillar morphology of typically 7–13 nm in diameter, a β-sheet secondary structure (known as cross-β) and ability to be stained by particular dyes, such as Congo red.[1] In the human body, amyloids have been linked to the development of various diseases.[2] Pathogenic amyloids form when previously healthy proteins lose their normal structure and physiological functions (misfolding) and form fibrous deposits within and around cells. These protein misfolding and deposition processes disrupt the healthy function of tissues and organs.
Such amyloids have been associated with (but not necessarily as the cause of) more than 50[2][3] human diseases, known as amyloidosis, and may play a role in some neurodegenerative diseases.[2][4] Some of these diseases are mainly sporadic and only a few cases are familial. Others are only familial. Some are iatrogenic as they result from medical treatment. Prions are an infectious form of amyloids that can act as a template to convert other non-infectious forms.[5] Amyloids may also have normal biological functions; for example, in the formation of fimbriae in some genera of bacteria, transmission of epigenetic traits in fungi, as well as pigment deposition and hormone release in humans.[6]
Amyloids have been known to arise from many different proteins.[2][7] These polypeptide chains generally form β-sheet structures that aggregate into long fibers; however, identical polypeptides can fold into multiple distinct amyloid conformations.[8] The diversity of the conformations may have led to different forms of the prion diseases.[6]
An unusual secondary structure named α sheet has been proposed as the toxic constituent of amyloid precursor proteins,[9] but this idea is not widely accepted at present.
Definition
The name amyloid comes from the early mistaken identification by Rudolf Virchow of the substance as starch (amylum in Latin, from Ancient Greek:), based on crude iodine-staining techniques. For a period, the scientific community debated whether or not amyloid deposits are fatty deposits or carbohydrate deposits until it was finally found (in 1859) that they are, in fact, deposits of albumoid proteinaceous material.[10]
- The classical, histopathological definition of amyloid is an extracellular, proteinaceous fibrillar deposit exhibiting β-sheet secondary structure and identified by apple-green birefringence when stained with congo red under polarized light. These deposits often recruit various sugars and other components such as serum amyloid P component, resulting in complex, and sometimes inhomogeneous structures.[11] Recently this definition has come into question as some classic, amyloid species have been observed in distinctly intracellular locations.[12]
- A more recent, biophysical definition is broader, including any polypeptide that polymerizes to form a cross-β structure, in vivo or in vitro, inside or outside cells. Microbiologists, biochemists, biophysicists, chemists and physicists have largely adopted this definition,[13][14] leading to some conflict in the biological community over an issue of language.
Proteins forming amyloids in diseases
To date, 37 human proteins have been found to form amyloid in pathology and be associated with well-defined diseases.[2] The International Society of Amyloidosis classifies amyloid fibrils and their associated diseases based upon associated proteins (for example ATTR is the group of diseases and associated fibrils formed by TTR).[3] A table is included below.
| Protein | Diseases | Official abbreviation |
|---|---|---|
| β amyloid peptide (Aβ) from Amyloid precursor protein[15][16][17][18] | Alzheimer's disease, Hereditary cerebral haemorrhage with amyloidosis | Aβ |
| α-synuclein[16] | Parkinson's disease, Parkinson's disease dementia, Dementia with Lewy bodies, Multiple system atrophy | AαSyn |
| PrPSc[19] | Transmissible spongiform encephalopathy (e.g. Fatal familial insomnia, Gerstmann-Sträussler-Scheinker disease, Creutzfeldt-Jacob disease, New variant Creutzfeldt-Jacob disease) | APrP |
| Microtubule-associated protein tau | Various forms of tauopathies (e.g. Pick's disease, Progressive supranuclear palsy, Corticobasal degeneration, Frontotemporal dementia with parkinsonism linked to chromosome 17, Argyrophilic grain disease) | ATau |
| Huntingtin exon 1[20][21] | Huntington's disease | HTTex1 |
| ABri peptide | Familial British dementia | ABri |
| ADan peptide | Familial Danish dementia | ADan |
| Fragments of immunoglobulin light chains[22] | Light chain amyloidosis | AL |
| Fragments of immunoglobulin heavy chains[22] | Heavy chain amyloidosis | AH |
| full length of N-terminal fragments of Serum amyloid A protein | AA amyloidosis | AA |
| Transthyretin | Senile systemic amyloidosis, Familial amyloid polyneuropathy, Familial amyloid cardiomyopathy, Leptomeningeal amyloidosis | ATTR |
| β-2 microglobulin | Dialysis related amyloidosis, Hereditary visceral amyloidosis (familial) | Aβ2M |
| N-terminal fragments of Apolipoprotein AI | ApoAI amyloidosis | AApoAI |
| C-terminally extended Apolipoprotein AII | ApoAII amyloidosis | AApoAII |
| N-terminal fragments of Apolipoprotein AIV | ApoAIV amyloidosis | AApoAIV |
| Apolipoprotein C-II | ApoCII amyloidosis | AApoCII |
| Apolipoprotein C-III | ApoCIII amyloidosis | AApoCIII |
| fragments of Gelsolin | Familial amyloidosis, Finnish type | AGel |
| Lysozyme | Hereditary non-neuropathic systemic amyloidosis | ALys |
| fragments of Fibrinogen α chain | Fibrinogen amyloidosis | AFib |
| N-terminally truncated Cystatin C | Hereditary cerebral hemorrhage with amyloidosis, Icelandic type | ACys |
| IAPP (Amylin)[23][24] | Diabetes mellitus type 2, Insulinoma | AIAPP |
| Calcitonin[22] | Medullary carcinoma of the thyroid | ACal |
| Atrial natriuretic factor | Cardiac arrhythmias, Isolated atrial amyloidosis | AANF |
| Prolactin | Pituitary prolactinoma | APro |
| Insulin | Injection-localized amyloidosis | AIns |
| Lactadherin / Medin | Aortic medial amyloidosis | AMed |
| Lactotransferrin / Lactoferrin | Gelatinous drop-like corneal dystrophy | ALac |
| Odontogenic ameloblast-associated protein | Calcifying epithelial odontogenic tumors | AOAAP |
| Pulmonary surfactant-associated protein C (SP-C) | Pulmonary alveolar proteinosis | ASPC |
| Leukocyte cell-derived chemotaxin-2 (LECT-2) | Renal LECT2 amyloidosis | ALECT2 |
| Galectin-7 | Lichen amyloidosis, Macular amyloidosis | AGal7 |
| Corneodesmosin | Hypotrichosis simplex of the scalp | ACor |
| C-terminal fragments of TGFBI/Keratoepithelin | Lattice corneal dystrophy type I, Lattice corneal dystrophy type 3A, Lattice corneal dystrophy Avellino type | AKer |
| Semenogelin-1 (SGI) | Seminal vesicle amyloidosis | ASem1 |
| Proteins S100A8/A9 | Prostate cancer | none |
| Enfuvirtide | Injection-localized amyloidosis | AEnf |
Non-disease and functional amyloids
Many examples of non-pathological amyloid with a well-defined physiological role have been identified in various organisms, including human. These may be termed as functional or physiological or native amyloid.[25][26][2]
- Functional amyloid in Homo sapiens:
- Functional amyloid in other organisms:
- Curli fibrils produced by E. coli, Salmonella, and a few other members of the Enterobacteriales (Csg). The genetic elements (operons) encoding the curli system are phylogenetic widespread and can be found in at least four bacterial phyla.[31] This suggest that many more bacteria may express curli fibrils.
- GvpA, forming the walls of particular Gas vesicles, i.e. the buoyancy organelles of aquatic archaea and eubacteria[32]
- Fap fibrils in various species of Pseudomonas[33][34]
- Chaplins from Streptomyces coelicolor[35]
- Spidroin from Trichonephila edulis (spider) (Spider silk)[36]
- Hydrophobins from Neurospora crassa and other fungi[37]
- Fungal cell adhesion proteins forming cell surface amyloid regions with greatly increased binding strength[38][39]
- Environmental biofilms according to staining with amyloid specific dyes and antibodies.[40]
- Tubular sheaths encasing Methanosaeta thermophila filaments[41]
- Functional amyloid acting as prions
- Several yeast prions are based on an infectious amyloid, e.g. [PSI+] (Sup35p); [URE3] (Ure2p); [PIN+] or [RNQ+] (Rnq1p); [SWI1+] (Swi1p) and [OCT8+] (Cyc8p)
- Prion HET-s from Podospora anserina[42]
- Neuron-specific isoform of CPEB from Aplysia californica (marine snail)[43]
Structure
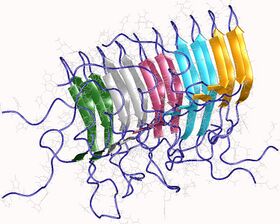
Amyloids are formed of long unbranched fibers that are characterized by an extended β-sheet secondary structure in which individual β strands (β-strands) (coloured arrows in the adjacent figure) are arranged in an orientation perpendicular to the long axis of the fiber. Such a structure is known as cross-β structure. Each individual fiber may be 7–13 nanometres in width and a few micrometres in length.[6][2] The main hallmarks recognised by different disciplines to classify protein aggregates as amyloid is the presence of a fibrillar morphology with the expected diameter, detected using transmission electron microscopy (TEM) or atomic force microscopy (AFM), the presence of a cross-β secondary structure, determined with circular dichroism, FTIR, solid-state nuclear magnetic resonance (ssNMR), X-ray crystallography, or X-ray fiber diffraction (often considered the "gold-standard" test to see whether a structure contains cross-β fibres), and an ability to stain with specific dyes, such as Congo red, thioflavin T or thioflavin S.[2]
The term "cross-β" was based on the observation of two sets of diffraction lines, one longitudinal and one transverse, that form a characteristic "cross" pattern.[45] There are two characteristic scattering diffraction signals produced at 4.7 and 10 Å (0.47 nm and 1.0 nm), corresponding to the interstrand and stacking distances in β sheets.[1] The "stacks" of β sheet are short and traverse the breadth of the amyloid fibril; the length of the amyloid fibril is built by aligned β-strands. The cross-β pattern is considered a diagnostic hallmark of amyloid structure.[6]
Amyloid fibrils are generally composed of 1–8 protofilaments (one protofilament also corresponding to a fibril is shown in the figure), each 2–7 nm in diameter, that interact laterally as flat ribbons that maintain the height of 2–7 nm (that of a single protofilament) and are up to 30 nm wide; more often protofilaments twist around each other to form the typically 7–13 nm wide fibrils.[2] Each protofilament possesses the typical cross-β structure and may be formed by 1–6 β-sheets (six are shown in the figure) stacked on each other. Each individual protein molecule can contribute one to several β-strands in each protofilament and the strands can be arranged in antiparallel β-sheets, but more often in parallel β-sheets. Only a fraction of the polypeptide chain is in a β-strand conformation in the fibrils, the remainder forms structured or unstructured loops or tails.
For a long time our knowledge of the atomic-level structure of amyloid fibrils was limited by the fact that they are unsuitable for the most traditional methods for studying protein structures. Recent years have seen progress in experimental methods, including solid-state NMR spectroscopy and Cryo-Electron Microscopy. Combined, these methods have provided 3D atomic structures of amyloid fibrils formed by amyloid β peptides, α-synuclein, tau, and the FUS protein, associated with various neurodegenerative diseases.[46][47]
X-ray diffraction studies of microcrystals revealed atomistic details of core region of amyloid, although only for simplified peptides having a length remarkably shorter than that of peptides or proteins involved in disease.[48][49] The crystallographic structures show that short stretches from amyloid-prone regions of amyloidogenic proteins run perpendicular to the filament axis, consistent with the "cross-β" feature of amyloid structure. They also reveal a number of characteristics of amyloid structures – neighboring β-sheets are tightly packed together via an interface devoid of water (therefore referred to as dry interface), with the opposing β-strands slightly offset from each other such that their side-chains interdigitate. This compact dehydrated interface created was termed a steric-zipper interface.[6] There are eight theoretical classes of steric-zipper interfaces, dictated by the directionality of the β-sheets (parallel and anti-parallel) and symmetry between adjacent β-sheets. A limitation of X-ray crystallography for solving amyloid structure is represented by the need to form microcrystals, which can be achieved only with peptides shorter than those associated with disease.
Although bona fide amyloid structures always are based on intermolecular β-sheets, different types of "higher order" tertiary folds have been observed or proposed. The β-sheets may form a β-sandwich, or a β-solenoid which may be either β-helix or β-roll. Native-like amyloid fibrils in which native β-sheet containing proteins maintain their native-like structure in the fibrils have also been proposed.[50] There are few developed ideas on how the complex backbone topologies of disulfide-constrained proteins, which are prone to form amyloid fibrils (such as insulin and lysozyme), adopt the amyloid β-sheet motif. The presence of multiple constraints significantly reduces the accessible conformational space, making computational simulations of amyloid structures more feasible. [51]
One complicating factor in studies of amyloidogenic polypeptides is that identical polypeptides can fold into multiple distinct amyloid conformations.[6] This phenomenon is typically described as amyloid polymorphism.[8][52] [53] It has notable biological consequences given that it is thought to explain the prion strain phenomenon.
Formation
File:Three phases of amyloid fibril formation.tif Amyloid is formed through the polymerization of hundreds to thousands of monomeric peptides or proteins into long fibers. Amyloid formation involves a lag phase (also called nucleation phase), an exponential phase (also called growth phase) and a plateau phase (also called saturation phase), as shown in the figure.[54][55][56][57] Indeed, when the quantity of fibrils is plotted versus time, a sigmoidal time course is observed reflecting the three distinct phases.
In the simplest model of 'nucleated polymerization' (marked by red arrows in the figure below), individual unfolded or partially unfolded polypeptide chains (monomers) convert into a nucleus (monomer or oligomer) via a thermodynamically unfavourable process that occurs early in the lag phase.[56] Fibrils grow subsequently from these nuclei through the addition of monomers in the exponential phase.[56]
A different model, called 'nucleated conformational conversion' and marked by blue arrows in the figure below, was introduced later on to fit some experimental observations: monomers have often been found to convert rapidly into misfolded and highly disorganized oligomers distinct from nuclei.[58] Only later on, will these aggregates reorganise structurally into nuclei, on which other disorganised oligomers will add and reorganise through a templating or induced-fit mechanism (this 'nucleated conformational conversion' model), eventually forming fibrils.[58]
Normally folded proteins have to unfold partially before aggregation can take place through one of these mechanisms.[59] In some cases, however, folded proteins can aggregate without crossing the major energy barrier for unfolding, by populating native-like conformations as a consequence of thermal fluctuations, ligand release or local unfolding occurring in particular circumstances.[59] In these native-like conformations, segments that are normally buried or structured in the fully folded and possessing a high propensity to aggregate become exposed to the solvent or flexible, allowing the formation of native-like aggregates, which convert subsequently into nuclei and fibrils. This process is called 'native-like aggregation' (green arrows in the figure) and is similar to the 'nucleated conformational conversion' model.
A more recent, modern and thorough model of amyloid fibril formation involves the intervention of secondary events, such as 'fragmentation', in which a fibril breaks into two or more shorter fibrils, and 'secondary nucleation', in which fibril surfaces (not fibril ends) catalyze the formation of new nuclei.[57] Both secondary events increase the number of fibril ends able to recruit new monomers or oligomers, therefore accelerating fibril formation through a positive feedback mechanism. These events add to the well recognised steps of primary nucleation (formation of the nucleus from the monomers through one of models described above), fibril elongation (addition of monomers or oligomers to growing fibril ends) and dissociation (opposite process).
Such a new model is described in the figure on the right and involves the utilization of a master equation that includes all steps of amyloid fibril formation, i.e. primary nucleation, fibril elongation, secondary nucleation and fibril fragmentation.[57][60] The rate constants of the various steps can be determined from a global fit of a number of time courses of aggregation (for example ThT fluorescence emission versus time) recorded at different protein concentrations.[57] The general master equation approach to amyloid fibril formation with secondary pathways has been developed by Knowles, Vendruscolo, Cohen, Michaels and coworkers and considers the time evolution of the concentration of fibrils of length (here represents the number of monomers in an aggregate).[60] where denotes the Kronecker delta. The physical interpretation of the various terms in the above master equation is straight forward: the terms on the first line describe the growth of fibrils via monomer addition with rate constant (elongation). The terms on the second line describe monomer dissociation, i.e. the inverse process of elongation. is the rate constant of monomer dissociation. The terms on the third line describe the effect of fragmentation, which is assumed to occur homogeneously along fibrils with rate constant . Finally, the terms on the last line describe primary and secondary nucleation respectively. Note that the rate of secondary nucleation is proportional to the mass of aggregates, defined as .
Following this analytical approach, it has become apparent that the lag phase does not correspond necessarily to only nucleus formation, but rather results from a combination of various steps. Similarly, the exponential phase is not only fibril elongation, but results from a combination of various steps, involving primary nucleation, fibril elongation, but also secondary events. A significant quantity of fibrils resulting from primary nucleation and fibril elongation may be formed during the lag phase and secondary steps, rather than only fibril elongation, can be the dominant processes contributing to fibril growth during the exponential phase. With this new model, any perturbing agents of amyloid fibril formation, such as putative drugs, metabolites, mutations, chaperones, etc., can be assigned to a specific step of fibril formation.
Amino acid sequence and amyloid formation
In general, amyloid polymerization (aggregation or non-covalent polymerization) is sequence-sensitive, that is mutations in the sequence can induce or prevent self-assembly.[61][62] For example, humans produce amylin, an amyloidogenic peptide associated with type II diabetes, but in rats and mice prolines are substituted in critical locations and amyloidogenesis does not occur.[citation needed] Studies comparing synthetic to recombinant β amyloid peptide in assays measuring rate of fibrillation, fibril homogeneity, and cellular toxicity showed that recombinant β amyloid peptide has a faster fibrillation rate and greater toxicity than synthetic β amyloid peptide.[63]
There are multiple classes of amyloid-forming polypeptide sequences.[8][52][53] Glutamine-rich polypeptides are important in the amyloidogenesis of Yeast and mammalian prions, as well as trinucleotide repeat disorders including Huntington's disease. When glutamine-rich polypeptides are in a β-sheet conformation, glutamines can brace the structure by forming inter-strand hydrogen bonding between its amide carbonyls and nitrogens of both the backbone and side chains. The onset age for Huntington's disease shows an inverse correlation with the length of the polyglutamine sequence, with analogous findings in a C. elegans model system with engineered polyglutamine peptides.[64]
Other polypeptides and proteins such as amylin and the β amyloid peptide do not have a simple consensus sequence and are thought to aggregate through the sequence segments enriched with hydrophobic residues, or residues with high propensity to form β-sheet structure.[61] Among the hydrophobic residues, aromatic amino-acids are found to have the highest amyloidogenic propensity.[65][66]
Cross-polymerization (fibrils of one polypeptide sequence causing other fibrils of another sequence to form) is observed in vitro and possibly in vivo. This phenomenon is important, since it would explain interspecies prion propagation and differential rates of prion propagation, as well as a statistical link between Alzheimer's and type 2 diabetes.[67] In general, the more similar the peptide sequence the more efficient cross-polymerization is, though entirely dissimilar sequences can cross-polymerize and highly similar sequences can even be "blockers" that prevent polymerization.[citation needed]
Amyloid toxicity
The reasons why amyloid cause diseases are unclear. In some cases, the deposits physically disrupt tissue architecture, suggesting disruption of function by some bulk process. An emerging consensus implicates prefibrillar intermediates, rather than mature amyloid fibers, in causing cell death, particularly in neurodegenerative diseases.[17][68] The fibrils are, however, far from innocuous, as they keep the protein homeostasis network engaged, release oligomers, cause the formation of toxic oligomers via secondary nucleation, grow indefinitely spreading from district to district[2] and, in some cases, may be toxic themselves.[69]
Calcium dysregulation has been observed to occur early in cells exposed to protein oligomers. These small aggregates can form ion channels through lipid bilayer membranes and activate NMDA and AMPA receptors. Channel formation has been hypothesized to account for calcium dysregulation and mitochondrial dysfunction by allowing indiscriminate leakage of ions across cell membranes.[70] Studies have shown that amyloid deposition is associated with mitochondrial dysfunction and a resulting generation of reactive oxygen species (ROS), which can initiate a signalling pathway leading to apoptosis.[71] There are reports that indicate amyloid polymers (such as those of huntingtin, associated with Huntington's disease) can induce the polymerization of essential amyloidogenic proteins, which should be deleterious to cells. Also, interaction partners of these essential proteins can also be sequestered.[72]
All these mechanisms of toxicity are likely to play a role. In fact, the aggregation of a protein generates a variety of aggregates, all of which are likely to be toxic to some degree. A wide variety of biochemical, physiological and cytological perturbations has been identified following the exposure of cells and animals to such species, independently of their identity. The oligomers have also been reported to interact with a variety of molecular targets. Hence, it is unlikely that there is a unique mechanism of toxicity or a unique cascade of cellular events. The misfolded nature of protein aggregates causes a multitude of aberrant interactions with a multitude of cellular components, including membranes, protein receptors, soluble proteins, RNAs, small metabolites, etc.
Histological staining
In the clinical setting, amyloid diseases are typically identified by a change in the spectroscopic properties of planar aromatic dyes such as thioflavin T, congo red or NIAD-4.[73] In general, this is attributed to the environmental change, as these dyes intercalate between β-strands to confine their structure.[74]
Congo Red positivity remains the gold standard for diagnosis of amyloidosis. In general, binding of Congo Red to amyloid plaques produces a typical apple-green birefringence when viewed under cross-polarized light. Recently, significant enhancement of fluorescence quantum yield of NIAD-4 was exploited to super-resolution fluorescence imaging of amyloid fibrils[75] and oligomers.[76] To avoid nonspecific staining, other histology stains, such as the hematoxylin and eosin stain, are used to quench the dyes' activity in other places such as the nucleus, where the dye might bind. Modern antibody technology and immunohistochemistry has made specific staining easier, but often this can cause trouble because epitopes can be concealed in the amyloid fold; in general, an amyloid protein structure is a different conformation from the one that the antibody recognizes.
See also
References
- ↑ 1.0 1.1 "Common core structure of amyloid fibrils by synchrotron X-ray diffraction". Journal of Molecular Biology 273 (3): 729–39. October 1997. doi:10.1006/jmbi.1997.1348. PMID 9356260.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 "Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade". Annual Review of Biochemistry 86: 27–68. June 2017. doi:10.1146/annurev-biochem-061516-045115. PMID 28498720.
- ↑ 3.0 3.1 "Amyloid nomenclature 2018: recommendations by the International Society of Amyloidosis (ISA) nomenclature committee". Amyloid 25 (4): 215–219. December 2018. doi:10.1080/13506129.2018.1549825. PMID 30614283.
- ↑ "Ubiquitous amyloids". Applied Biochemistry and Biotechnology 166 (7): 1626–43. April 2012. doi:10.1007/s12010-012-9549-3. PMID 22350870.
- ↑ "Amyloids, prions and the inherent infectious nature of misfolded protein aggregates". Trends in Biochemical Sciences 31 (3): 150–5. March 2006. doi:10.1016/j.tibs.2006.01.002. PMID 16473510.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 "Amyloid structure: conformational diversity and consequences". Annual Review of Biochemistry 80: 557–85. 2011. doi:10.1146/annurev-biochem-090908-120656. PMID 21456964.
- ↑ "A systematic exploration of the influence of the protein stability on amyloid fibril formation in vitro". Proceedings of the National Academy of Sciences of the United States of America 97 (16): 8979–84. August 2000. doi:10.1073/pnas.150091797. PMID 10908649. Bibcode: 2000PNAS...97.8979R.
- ↑ 8.0 8.1 8.2 "Amyloid fibril formation by Aβ16-22, a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR". Biochemistry 39 (45): 13748–59. November 2000. doi:10.1021/bi0011330. PMID 11076514.
- ↑ "Pauling and Coreys α-pleated sheet structure may define the prefibrillar amyloidogenic intermediate in amyloid disease". Proceedings of the National Academy of Sciences of the United States of America 101 (1): 11622–11627. 2004. doi:10.1073/pnas.0401781101. PMID 15280548. Bibcode: 2004PNAS..10111622A.
- ↑ "Amyloidosis: a convoluted story". British Journal of Haematology 114 (3): 529–38. September 2001. doi:10.1046/j.1365-2141.2001.02999.x. PMID 11552976.
- ↑ "Review: history of the amyloid fibril". Journal of Structural Biology 130 (2–3): 88–98. June 2000. doi:10.1006/jsbi.2000.4221. PMID 10940217.
- ↑ "Toxic human islet amyloid polypeptide (h-IAPP) oligomers are intracellular, and vaccination to induce anti-toxic oligomer antibodies does not prevent h-IAPP-induced β-cell apoptosis in h-IAPP transgenic mice". Diabetes 56 (5): 1324–32. May 2007. doi:10.2337/db06-1579. PMID 17353506.
- ↑ "Techniques to study amyloid fibril formation in vitro". Methods 34 (1): 151–60. September 2004. doi:10.1016/j.ymeth.2004.03.012. PMID 15283924.
- ↑ "On the structural definition of amyloid fibrils and other polypeptide aggregates". Cellular and Molecular Life Sciences 64 (16): 2066–78. August 2007. doi:10.1007/s00018-007-7110-2. PMID 17530168.
- ↑ "The many faces of amyloid β in Alzheimer's disease". Current Molecular Medicine 8 (6): 580–4. September 2008. doi:10.2174/156652408785747951. PMID 18781964.
- ↑ 16.0 16.1 "Protein aggregation in the brain: the molecular basis for Alzheimer's and Parkinson's diseases". Molecular Medicine 14 (7–8): 451–64. 2008. doi:10.2119/2007-00100.Irvine. PMID 18368143.
- ↑ 17.0 17.1 "Soluble protein oligomers as emerging toxins in Alzheimer's and other amyloid diseases". IUBMB Life 59 (4–5): 332–45. 2007. doi:10.1080/15216540701283882. PMID 17505973.
- ↑ "The amyloid β peptide: a chemist's perspective. Role in Alzheimer's and fibrillization". Chemical Reviews 112 (10): 5147–92. October 2012. doi:10.1021/cr3000994. PMID 22813427. http://centaur.reading.ac.uk/30230/2/AbetaRevisednew%20-IWH%20%281%29.pdf.
- ↑ "More than just mad cow disease". Nature Structural Biology 8 (4): 281. April 2001. doi:10.1038/86132. PMID 11276238.
- ↑ "Huntington's disease: revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases". The FEBS Journal 275 (17): 4252–62. September 2008. doi:10.1111/j.1742-4658.2008.06561.x. PMID 18637947.
- ↑ "Targeting protein aggregation in neurodegeneration--lessons from polyglutamine disorders". Expert Opinion on Therapeutic Targets 10 (4): 505–13. August 2006. doi:10.1517/14728222.10.4.505. PMID 16848688.
- ↑ 22.0 22.1 22.2 Amyloidosis: Definition of Amyloid and Amyloidosis, Classification Systems, Systemic Amyloidoses. 10 October 2018. https://emedicine.medscape.com/article/335414-overview.
- ↑ "Islet amyloid in type 2 diabetes, and the toxic oligomer hypothesis". Endocrine Reviews 29 (3): 303–16. May 2008. doi:10.1210/er.2007-0037. PMID 18314421.
- ↑ "Islet amyloid and type 2 diabetes mellitus". The New England Journal of Medicine 343 (6): 411–9. August 2000. doi:10.1056/NEJM200008103430607. PMID 10933741.
- ↑ "Amyloids: friend or foe?". Journal of Alzheimer's Disease 13 (4): 407–19. May 2008. doi:10.3233/JAD-2008-13406. PMID 18487849. PMC 2674399. http://iospress.metapress.com/openurl.asp?genre=article&issn=1387-2877&volume=13&issue=4&spage=407.
- ↑ "Functional amyloid--from bacteria to humans". Trends in Biochemical Sciences 32 (5): 217–24. May 2007. doi:10.1016/j.tibs.2007.03.003. PMID 17412596.
- ↑ "Functional amyloid formation within mammalian tissue". PLOS Biology 4 (1): e6. January 2006. doi:10.1371/journal.pbio.0040006. PMID 16300414.
- ↑ "Functional amyloids as natural storage of peptide hormones in pituitary secretory granules". Science 325 (5938): 328–32. July 2009. doi:10.1126/science.1173155. PMID 19541956. Bibcode: 2009Sci...325..328M.
- ↑ "The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis". Cell 150 (2): 339–50. July 2012. doi:10.1016/j.cell.2012.06.019. PMID 22817896.
- ↑ "Direct visualization of HIV-enhancing endogenous amyloid fibrils in human semen". Nature Communications 5: 3508. April 2014. doi:10.1038/ncomms4508. PMID 24691351. Bibcode: 2014NatCo...5.3508U.
- ↑ "Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure". PLOS ONE 7 (12): e51274. 2012. doi:10.1371/journal.pone.0051274. PMID 23251478. Bibcode: 2012PLoSO...751274D.
- ↑ "An amyloid organelle, solid-state NMR evidence for cross-β assembly of gas vesicles". The Journal of Biological Chemistry 287 (5): 3479–84. January 2012. doi:10.1074/jbc.M111.313049. PMID 22147705.
- ↑ "Functional amyloid in Pseudomonas". Molecular Microbiology 77 (4): 1009–20. August 2010. doi:10.1111/j.1365-2958.2010.07269.x. PMID 20572935.
- ↑ "Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation". MicrobiologyOpen 2 (3): 365–82. June 2013. doi:10.1002/mbo3.81. PMID 23504942.
- ↑ "A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils". Genes & Development 17 (14): 1714–26. July 2003. doi:10.1101/gad.264303. PMID 12832396.
- ↑ "Amyloidogenic nature of spider silk". European Journal of Biochemistry 269 (16): 4159–63. August 2002. doi:10.1046/j.1432-1033.2002.03112.x. PMID 12180993.
- ↑ "The hydrophobin EAS is largely unstructured in solution and functions by forming amyloid-like structures". Structure 9 (2): 83–91. February 2001. doi:10.1016/s0969-2126(00)00559-1. PMID 11250193.
- ↑ "A role for amyloid in cell aggregation and biofilm formation". PLOS ONE 6 (3): e17632. March 2011. doi:10.1371/journal.pone.0017632. PMID 21408122. Bibcode: 2011PLoSO...617632G.
- ↑ "Strengthening relationships: amyloids create adhesion nanodomains in yeasts". Trends in Microbiology 20 (2): 59–65. February 2012. doi:10.1016/j.tim.2011.10.002. PMID 22099004.
- ↑ "Amyloid adhesins are abundant in natural biofilms". Environmental Microbiology 9 (12): 3077–90. December 2007. doi:10.1111/j.1462-2920.2007.01418.x. PMID 17991035.
- ↑ "The Tubular Sheaths Encasing Methanosaeta thermophila Filaments Are Functional Amyloids". The Journal of Biological Chemistry 290 (33): 20590–600. August 2015. doi:10.1074/jbc.M115.654780. PMID 26109065.
- ↑ "The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog". Proceedings of the National Academy of Sciences of the United States of America 94 (18): 9773–8. September 1997. doi:10.1073/pnas.94.18.9773. PMID 9275200. Bibcode: 1997PNAS...94.9773C.
- ↑ "A neuronal isoform of the aplysia CPEB has prion-like properties". Cell 115 (7): 879–91. December 2003. doi:10.1016/s0092-8674(03)01020-1. PMID 14697205.
- ↑ "Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils". PNAS 105 (47): 18349–54. 25 November 2008. doi:10.1073/pnas.0806270105. PMID 19015532. Bibcode: 2008PNAS..10518349P.
- ↑ Wormell RL. New fibres from proteins. Academic Press, 1954, p. 106.
- ↑ "Emerging Structural Understanding of Amyloid Fibrils by Solid-State NMR". Trends in Biochemical Sciences 42 (10): 777–787. October 2017. doi:10.1016/j.tibs.2017.08.001. PMID 28916413.
- ↑ "Cryo-EM structures of tau filaments from Alzheimer's disease". Nature 547 (7662): 185–190. July 2017. doi:10.1038/nature23002. PMID 28678775. Bibcode: 2017Natur.547..185F.
- ↑ "Structure of the cross-β spine of amyloid-like fibrils". Nature 435 (7043): 773–8. June 2005. doi:10.1038/nature03680. PMID 15944695. Bibcode: 2005Natur.435..773N.
- ↑ "Atomic structures of amyloid cross-β spines reveal varied steric zippers". Nature 447 (7143): 453–7. May 2007. doi:10.1038/nature05695. PMID 17468747. Bibcode: 2007Natur.447..453S.
- ↑ "Arrangement of subunits and ordering of β-strands in an amyloid sheet". Nature Structural Biology 9 (10): 734–9. October 2002. doi:10.1038/nsb838. PMID 12219081.
- ↑ Puławski, W; Dzwolak, W (7 June 2022). "Virtual Quasi-2D Intermediates as Building Blocks for Plausible Structural Models of Amyloid Fibrils from Proteins with Complex Topologies: A Case Study of Insulin.". Langmuir 38 (22): 7024–7034. doi:10.1021/acs.langmuir.2c00699. PMID 35617668.
- ↑ 52.0 52.1 "Molecular alignment within β-sheets in Aβ14-23 fibrils: solid-state NMR experiments and theoretical predictions". Biophysical Journal 92 (2): 594–602. January 2007. doi:10.1529/biophysj.106.091017. PMID 17056725. Bibcode: 2007BpJ....92..594B.
- ↑ 53.0 53.1 "Assembling amyloid fibrils from designed structures containing a significant amyloid β-peptide fragment". The Biochemical Journal 366 (Pt 1): 343–51. August 2002. doi:10.1042/BJ20020229. PMID 12023906.
- ↑ "The carboxy terminus of the β amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer's disease". Biochemistry 32 (18): 4693–7. May 1993. doi:10.1021/bi00069a001. PMID 8490014.
- ↑ "Analysis of protein aggregation kinetics". Amyloid, Prions, and Other Protein Aggregates. Methods in Enzymology. 309. 1999. pp. 256–74. doi:10.1016/s0076-6879(99)09019-9. ISBN 9780121822101.
- ↑ 56.0 56.1 56.2 "Protein aggregation kinetics, mechanism, and curve-fitting: a review of the literature". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1794 (3): 375–97. March 2009. doi:10.1016/j.bbapap.2008.10.016. PMID 19071235.
- ↑ 57.0 57.1 57.2 57.3 "An analytical solution to the kinetics of breakable filament assembly". Science 326 (5959): 1533–7. December 2009. doi:10.1126/science.1178250. PMID 20007899. Bibcode: 2009Sci...326.1533K.
- ↑ 58.0 58.1 "Nucleated conformational conversion and the replication of conformational information by a prion determinant". Science 289 (5483): 1317–21. August 2000. doi:10.1126/science.289.5483.1317. PMID 10958771. Bibcode: 2000Sci...289.1317S.
- ↑ 59.0 59.1 "Amyloid formation by globular proteins under native conditions". Nature Chemical Biology 5 (1): 15–22. January 2009. doi:10.1038/nchembio.131. PMID 19088715.
- ↑ 60.0 60.1 "Chemical Kinetics for Bridging Molecular Mechanisms and Macroscopic Measurements of Amyloid Fibril Formation". Annual Review of Physical Chemistry 69 (1): 273–298. April 2018. doi:10.1146/annurev-physchem-050317-021322. PMID 29490200. Bibcode: 2018ARPC...69..273M.
- ↑ 61.0 61.1 "Rationalization of the effects of mutations on peptide and protein aggregation rates". Nature 424 (6950): 805–8. August 2003. doi:10.1038/nature01891. PMID 12917692. Bibcode: 2003Natur.424..805C.
- ↑ "Inhibition of amyloid fibril formation by peptide analogues modified with α-aminoisobutyric acid". Angewandte Chemie 43 (31): 4041–4. August 2004. doi:10.1002/anie.200353565. PMID 15300690.
- ↑ "The recombinant amyloid-β peptide Aβ1-42 aggregates faster and is more neurotoxic than synthetic Aβ-42". Journal of Molecular Biology 396 (1): 9–18. February 2010. doi:10.1016/j.jmb.2009.12.016. PMID 20026079.
- ↑ "The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans". Proceedings of the National Academy of Sciences of the United States of America 99 (16): 10417–22. August 2002. doi:10.1073/pnas.152161099. PMID 12122205. Bibcode: 2002PNAS...9910417M.
- ↑ "A possible role for pi-stacking in the self-assembly of amyloid fibrils". FASEB Journal 16 (1): 77–83. January 2002. doi:10.1096/fj.01-0442hyp. PMID 11772939.
- ↑ "Prediction of "aggregation-prone" and "aggregation-susceptible" regions in proteins associated with neurodegenerative diseases". Journal of Molecular Biology 350 (2): 379–92. July 2005. doi:10.1016/j.jmb.2005.04.016. PMID 15925383.
- ↑ "Amylin deposition in the brain: A second amyloid in Alzheimer disease?". Annals of Neurology 74 (4): 517–26. October 2013. doi:10.1002/ana.23956. PMID 23794448.
- ↑ "Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers". The Journal of Biological Chemistry 280 (17): 17294–300. April 2005. doi:10.1074/jbc.M500997200. PMID 15722360.
- ↑ "Unlike twins: an NMR comparison of two α-synuclein polymorphs featuring different toxicity". PLOS ONE 9 (3): e90659. March 5, 2014. doi:10.1371/journal.pone.0090659. PMID 24599158. Bibcode: 2014PLoSO...990659G.
- ↑ "Amyloid peptide channels". The Journal of Membrane Biology 202 (1): 1–10. November 2004. doi:10.1007/s00232-004-0709-4. PMID 15702375.
- ↑ "Amyloid β induces neuronal cell death through ROS-mediated ASK1 activation". Cell Death and Differentiation 12 (1): 19–24. January 2005. doi:10.1038/sj.cdd.4401528. PMID 15592360.
- ↑ "Amyloid-mediated sequestration of essential proteins contributes to mutant huntingtin toxicity in yeast". PLOS ONE 7 (1): e29832. 2012. doi:10.1371/journal.pone.0029832. PMID 22253794. Bibcode: 2012PLoSO...729832K.
- ↑ "In vivo optical imaging of amyloid aggregates in brain: design of fluorescent markers". Angewandte Chemie 44 (34): 5452–6. August 2005. doi:10.1002/anie.200500845. PMID 16059955.
- ↑ "Torsion-dependent fluorescence switching of amyloid-binding dye NIAD-4". Chemical Physics Letters 633: 109–13. 2015. doi:10.1016/j.cplett.2015.05.010. Bibcode: 2015CPL...633..109B.
- ↑ "Superresolution imaging of amyloid fibrils with binding-activated probes". ACS Chemical Neuroscience 4 (7): 1057–61. July 2013. doi:10.1021/cn400091m. PMID 23594172.
- ↑ "Morphological analysis of oligomeric vs. fibrillar forms of α-synuclein aggregates with super-resolution BALM imaging". Chemical Physics Letters 690: 62–67. 2017. doi:10.1016/j.cplett.2017.10.034. Bibcode: 2017CPL...690...62H.
External links
- Bacterial Inclusion Bodies Contain Amyloid-Like Structure at SciVee
- Amyloid Cascade Hypothesis
- Amyloid: Journal of Protein Folding Disorders web page
- Role of anesthetics in Alzheimer's disease: Molecular details revealed
 |
