Biology:Cdc6
| Cell division protein Cdc6/18 | |
|---|---|
| Identifiers | |
| Symbol | Cdc6 |
| InterPro | IPR016314 |
| Orc1/Cdc6-type DNA replication protein, archaea | |
|---|---|
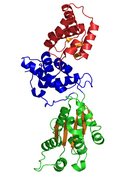 Crystal Structure of CDC6 from P. aerophilum (PDB: 1FNN). Green and blue domains are AAA domains; red domain is the Cdc6 C-terminal HTH domain (infobox below).[1] | |
| Identifiers | |
| Symbol | Orc1/Cdc6_arc |
| InterPro | IPR014277 |
| Cdc6/Orc1, C-terminal | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Cdc6_C | ||||||||
| Pfam | PF09079 | ||||||||
| InterPro | IPR015163 | ||||||||
| CDD | cd08768 | ||||||||
| |||||||||
Cdc6, or cell division cycle 6, is a protein in eukaryotic cells. It is mainly studied in the budding yeast Saccharomyces cerevisiae (P09119). It is an essential regulator of DNA replication and plays important roles in the activation and maintenance of the checkpoint mechanisms in the cell cycle that coordinate S phase and mitosis. It is part of the pre-replicative complex (pre-RC) and is required for loading minichromosome maintenance (MCM) proteins onto the DNA, an essential step in the initiation of DNA synthesis. In addition, it is a member of the family of AAA+ ATPases and highly related to ORC1; both are the same protein in archaea.[2]
Function
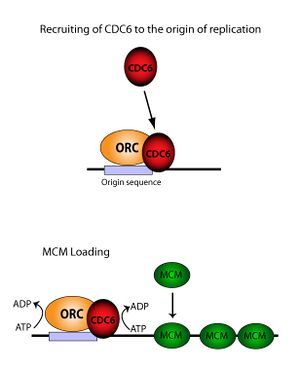
CDC6 is an ATP binding protein and a member of the pre-replicative complex (pre-RC) together with the origin recognition complex (ORC), Cdt1 and the MCM complex (containing MCM2-7p). CDC6 assembles after ORC in an ATP dependent manner and is required for loading MCM proteins onto the DNA. Reconstruction of electron microscope images showed that the ORC-CDC6 complex forms a ring-shaped structure with similar dimensions to those of ring-shaped MCM helicase.[4] A near-atomic resolution model of the entire ORC-Cdc6-Cdt1-Mcm2-7 (OCCM) complex with DNA was assembled from EM data in 2017.[5] It is thought that the CDC6-Cdt1 complex uses ATP hydrolysis to thread DNA through the central hole of the MCM doughnut.[6] Mutations in the binding motif of CDC6 strongly suggest that ATP binding and hydrolysis is essential for its function.[7] The minimal requirement for DNA binding has been mapped within its 47-amino acid sequence.[8] Furthermore, Cdc6 indirectly inhibits activation of the p34cdc2/CDC28 M phase kinase, thus nuclear division is suppressed.[9]
Regulation
CDC6 is normally present at high levels during the G1 phase of the cell cycle. This is partly because the CDC6 gene is only transcribed during G1 phase. On the onset of the S phase, CDC6 gets phosphorylated by the Cdc28-Clb5-Clb6 complex (Cdk2) and consequently becoming inactivated. This has been shown by introducing mutations in CDC6 at the consensus sites for Cdk2 phosphorylation (near the N-terminus) which inhibit degradation. The phosphorylation can furthermore be catalyzed by Cdc28-Cln. The inactivated CDC6 is then targeted for degradation by SCFCDC4-dependent ubiquitinylation and afterwards degraded by the proteasome. Thus, the regulation of CDC6 is tightly correlated to the activity of Cdk2 and since Cdk2-activity is oscillating once per cell cycle, the accumulation and degradation of CDC6 also oscillates.
Two states can be distinguished. In the first state (during G1 phase) Cdk2-activity is low, CDC6 can accumulate, hence the pre-RC can be formed but not activated. In the second state Cdk2-activity is high, CDC6 becomes inactivated, hence the pre-RC is activated but not formed. This change assures that DNA replication is performed only once per cell cycle. It has been shown that overexpression of CDC6 does not induce re-replication in cognate cells, probably due to inhibition through CDK that resets the cell cycle clock to G1. Nevertheless, it has been suggested that regulation of CDC6 is one of several redundant mechanisms that prevent re-replication of the DNA in eukaryotic cells.[10]
Structure
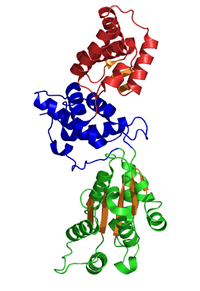
The crystallographic structure of a Cdc6/Orc1-related protein from the archaeon Pyrobaculum aerophilum has been solved and three structural domains have been identified.[1] Domain I and II form the ATP binding/hydrolysis site and are similar to other AAA+ ATPases. Domain III is structurally related to a winged-helix domain, thus may interact with origin DNA. From studies with E. coli γ clamp loading complex, it was suggested that domain III mediates protein-protein interactions with other AAA+ ATPases in the pre-RC, thus suggesting that the CDC6 builds a homodimer in its native form. The domains I and II form a cashew-shaped molecule that bind ATP in the cleft and additionally build the sensor motif for ATP/ADP recognition. These domains are also thought to mediate subsequent conformational changes. Nevertheless, the exact functional roles of these domains remain unclear.[8]
Disease
It has been shown CDC6 shows proto-oncogenic activity. Cdc6 overexpression interferes with the expression of INK4/ARF tumor suppressor genes through a mechanism involving the epigenetic modification of chromatin at the INK4/ARF locus. In addition, CDC6 overexpression in primary cells may promote DNA hyper-replication and induce a senescence response similar to that caused by oncogene activation. These findings indicate that deregulation of CDC6 expression in human cells poses a serious risk of carcinogenesis.[3] Down-regulation of CDC6 in prostate cancer was observed and associated with phenotypic characteristics of aggressive prostate cancer.[11] Furthermore, it has been observed that Cdc6 is greatly up-regulated in cervical cancer, lung cancer and brain cancer.[12]
See also
References
- ↑ 1.0 1.1 "Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control". Molecular Cell 6 (3): 637–648. September 2000. doi:10.1016/S1097-2765(00)00062-9. PMID 11030343.
- ↑ "Diversity of DNA Replication in the Archaea". Genes 8 (2): 56. January 2017. doi:10.3390/genes8020056. PMID 28146124.
- ↑ 3.0 3.1 "CDC6: from DNA replication to cell cycle checkpoints and oncogenesis". Carcinogenesis 29 (2): 237–243. February 2008. doi:10.1093/carcin/bgm268. PMID 18048387.
- ↑ "ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA". Nature Structural & Molecular Biology 12 (11): 965–971. November 2005. doi:10.1038/nsmb1002. PMID 16228006.
- ↑ "Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1". Nature Structural & Molecular Biology 24 (3): 316–324. March 2017. doi:10.1038/nsmb.3372. PMID 28191893.
- ↑ Cell biology. Saunders Elsevier. 2007. pp. 766–767. ISBN 978-1-4160-2255-8.
- ↑ "DNA replication in eukaryotic cells". Annual Review of Biochemistry 71: 333–374. 2002. doi:10.1146/annurev.biochem.71.110601.135425. PMID 12045100.
- ↑ 8.0 8.1 "Identification and characterization of Saccharomyces cerevisiae Cdc6 DNA-binding properties". Molecular Biology of the Cell 11 (5): 1673–1685. May 2000. doi:10.1091/mbc.11.5.1673. PMID 10793143.
- ↑ "Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division". The EMBO Journal 11 (6): 2167–2176. June 1992. doi:10.1002/j.1460-2075.1992.tb05276.x. PMID 1600944.
- ↑ "The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast". The EMBO Journal 16 (19): 5966–5976. October 1997. doi:10.1093/emboj/16.19.5966. PMID 9312054.
- ↑ "Down-regulation of Cdc6, a cell cycle regulatory gene, in prostate cancer". The Journal of Biological Chemistry 277 (28): 25431–25438. July 2002. doi:10.1074/jbc.M201199200. PMID 12006585.
- ↑ "The role of pre-replicative complex (pre-RC) components in oncogenesis". FASEB Journal 21 (14): 3786–3794. December 2007. doi:10.1096/fj.07-8900rev. PMID 17690155.
External links
 |

