Biology:Histone octamer
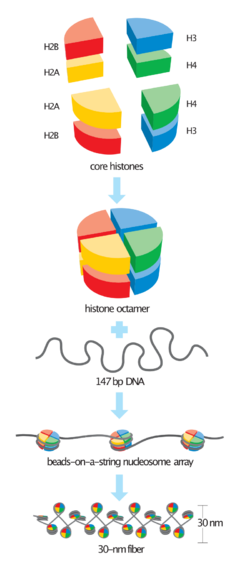
In molecular biology, a histone octamer is the eight-protein complex found at the center of a nucleosome core particle. It consists of two copies of each of the four core histone proteins (H2A, H2B, H3, and H4). The octamer assembles when a tetramer, containing two copies of H3 and two of H4, complexes with two H2A/H2B dimers. Each histone has both an N-terminal tail and a C-terminal histone-fold. Each of these key components interacts with DNA in its own way through a series of weak interactions, including hydrogen bonds and salt bridges. These interactions keep the DNA and the histone octamer loosely associated, and ultimately allow the two to re-position or to separate entirely.
History of research
Histone post-translational modifications were first identified and listed as having a potential regulatory role on the synthesis of RNA in 1964.[1] Since then, over several decades, chromatin theory has evolved. Chromatin subunit models as well as the notion of the nucleosome were established in 1973 and 1974, respectively.[2] Richmond and his research group has been able to elucidate the crystal structure of the histone octamer with DNA wrapped up around it at a resolution of 7 Å in 1984.[3] The structure of the octameric core complex was revisited seven years later and a resolution of 3.1 Å was elucidated for its crystal at a high salt concentration. Though sequence similarity is low between the core histones, each of the four have a repeated element consisting of a helix-loop-helix called the histone fold motif.[4] Furthermore, the details of protein-protein and protein-DNA interactions were fine-tuned by X-ray crystallography studies at 2.8 and 1.9 Å, respectively, in the 2000s.[5]
The histone octamer in molecular detail
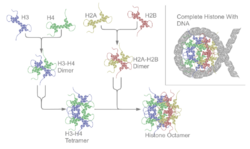
Core histones are four proteins called H2A, H2B, H3 and H4 and they are all found in equal parts in the cell. All four of the core histone amino acid sequences contain between 20 and 24% of lysine and arginine and the size or the protein ranges between 11400 and 15400 daltons, making them relatively small, yet highly positively charged proteins.[6] High content of positively charged amino acids allow them to closely associate with negatively charged DNA. Heterodimers, or histone-only intermediates are formed from histone-fold domains. The formation of histone only-intermediates proceeds when core histones are paired into the interlocked crescent shape quasi-symmetric heterodimer. Each histone fold domain is composed of 3 α-helix regions that are separated by disordered loops. The histone fold domain is responsible for formation of head-to-tail heterodimers of two histones: H2A-H2B and H3-H4. However, H3 and H4 histones first form a heterodimer and then in turn the heterodimer dimerizes to form a tetramer (H3-H4)2.[7] The heterodimer formation is based on the interaction of hydrophobic amino acid residue interactions between the two proteins.[7]
Quasi symmetry allows the heterodimer to be superimposed on itself by a 180 degree rotation around this symmetry axis. As a result of the rotation, two ends of histones involved in DNA binding of the crescent shape H3-H4 are equivalent, yet they organize different stretches of DNA. The H2A-H2B dimer also folds similarly. The (H3-H4)2 tetramer is wrapped with DNA around it as a first step of nucleosome formation. Then two H2A-H2B dimers are connected to the DNA-(H3-H4)2 complex to form a nucleosome.[8]
Each of the four core histones, in addition to their histone-fold domains, also contain flexible, unstructured extensions called histone “tails”.[9] Treatment of nucleosomes with protease trypsin indicates that after histone tails are removed, DNA is able to stay tightly bound to the nucleosome.[6] Histone tails are subject to a wide array of modifications which includes phosphorylation, acetylation, and methylation of serine, lysine and arginine residues.[6]
The histone octamer in the nucleosome
The nucleosome core particle is the most basic form of DNA compaction in eukaryotes. Nucleosomes consist of a histone octamer surrounded by 146 base pairs of DNA wrapped in a superhelical manner.[10] In addition to compacting the DNA, the histone octamer plays a key role in the transcription of the DNA surrounding it. The histone octamer interacts with the DNA through both its core histone folds and N-terminal tails. The histone fold interacts chemically and physically with the DNA's minor groove. Studies have found that the histones interact more favorably with A:T enriched regions than G:C enriched regions in the minor grooves.[6] The N-terminal tails do not interact with a specific region of DNA but rather stabilize and guide the DNA wrapped around the octamer. The interactions between the histone octamer and DNA, however, are not permanent. The two can be separated quite easily and often are during replication and transcription. Specific remodeling proteins are constantly altering the chromatin structure by breaking the bonds between the DNA and nucleosome.
Histone/DNA interactions
Histones are composed of mostly positively charged amino acid residues such as lysine and arginine. The positive charges allow them to closely associate with the negatively charged DNA through electrostatic interactions. Neutralizing the charges in the DNA allows it to become more tightly packed.[6]
Interactions with the minor groove
The histone-fold domains’ interaction with the minor groove accounts for the majority of the interactions in the nucleosome.[11] As the DNA wraps around the histone octamer, it exposes its minor groove to the histone octamer at 14 distinct locations. At these sites, the two interact through a series of weak, non-covalent bonds. The main source of bonds comes from hydrogen bonds, both direct and water-mediated.[10] The histone-fold hydrogen bonds with both phosphodiester backbone and the A:T rich bases. In these interactions, the histone fold binds to the oxygen atoms and hydroxyl side chains, respectively.[11] Together these sites have a total of about 40 hydrogen bonds, most of which are from the backbone interactions.[6] Additionially, 10 out of the 14 times that the minor groove faces the histone fold, an arginine side chain from the histone fold is inserted into the minor groove. The other four times, the arginine comes from a tail region of the histone.[11]
Tail interactions and modifications
As mentioned above the histone tails have been shown to directly interact with the DNA of the nucleosome. Each histone in the octamer has an N-terminal tail that protrudes from the histone core. The tails play roles both in inter and intra nucleosomal interactions that ultimately influence gene access.[12] Histones are positively charged molecules which allow a tighter bonding to the negatively charged DNA molecule. Reducing the positive charge of histone proteins reduces the strength of binding between the histone and DNA, making it more open to gene transcription (expression).[12] Moreover, these flexible units direct DNA wrapping in a left-handed manner around the histone octamer during nucleosome formation.[6] Once the DNA is bound the tails continue to interact with the DNA. The parts of the tail closest to the DNA hydrogen bond and strengthen the DNA's association with the octamer; the parts of the tail furthest away from the DNA, however, work in a very different manner. Cellular enzymes modify the amino acids in the distal sections of the tail to influence the accessibility of the DNA. The tails have also been implicated in the stabilization of 30-nm fibers. Research has shown removing certain tails prevents the nucleosomes from forming properly and a general failure to produce chromatin fiber.[12] In all, these associations protect the nucleosomal DNA from the external environment but also lower their accessibility to cellular replication and transcriptional machinery.
Nucleosome remodeling and disassembly
In order to access the nucleosomal DNA, the bonds between it and the histone octamer must be broken. This change takes place periodically in the cell as specific regions are transcribed, and it happens genome-wide during replication. Remodeling proteins work in three distinct ways: they can slide the DNA along the surface of the octamer, replace the one histone dimer with a variant, or remove the histone octamer entirely. No matter the method, in order to modify the nucleosomes, the remodeling complexes require energy from ATP hydrolysis to drive their actions.
Of the three techniques, sliding is the most common and least extreme.[13] The basic premise of the technique is to free up a region of DNA that the histone octamer would normally tightly bind. While the technique is not well defined, the most prominent hypothesis is that the sliding is done in an “inchworm” fashion. In this method, using ATP as an energy source, the translocase domain of the nucleosome-remodeling complex detaches a small region of DNA from the histone octamer. This “wave” of DNA, spontaneously breaking and remaking the hydrogen bonds as it goes, then propagates down the nucleosomal DNA until it reaches the last binding site with the histone octamer. Once the wave reaches the end of the histone octamer the excess that was once at the edge is extended into the region of linker DNA. In total, one round of this method moves the histone octamer several base pairs in a particular direction—away from the direction the “wave” propagated.[6][14]
Clinical relevance
Numerous reports show a link between age-related diseases, birth defects, and several types of cancer with disruption of certain histone post translational modifications. Studies have identified that N- and C-terminal tails are main targets for acetylation, methylation, ubiquitination and phosphorylation.[15] New evidence is pointing to several modifications within the histone core. Research is turning towards deciphering the role of these histone core modifications at the histone-DNA interface in the chromatin. p300 and cAMP response element-binding protein (CBP) possess histone acetyltransferase activity. p300 and CBP are the most promiscuous histone acetyltransferase enzymes acetylating all four core histones on multiple residues.[16] Lysine 18 and Lysine 27 on H3 were the only histone acetylation sites reduced upon CBP and p300 depletion in mouse embryonic fibroblasts.[17] Also, CBP and p300 knockout mice have an open neural tube defect and therefore die before birth. p300−/− embryos exhibit defective development of the heart. CBP+/− mice display growth retardation, craniofacial abnormalities, hematological malignancies, which are not observed in mice with p300+/−.[18] Mutations of both p300 have been reported in human tumors such as colorectal, gastric, breast, ovarian, lung, and pancreatic carcinomas. Also, activation or localization of two histone acetyltransferases can be oncogenic.
See also
References
- ↑ Allfrey, VG; Mirsky, AE (May 1, 1964). "Structural Modifications of Histones and their Possible Role in the Regulation of RNA Synthesis.". Science 144 (3618): 559. doi:10.1126/science.144.3618.559. PMID 17836360.
- ↑ Hewish, Dean R.; Burgoyne, Leigh A. (1973). "Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease". Biochem. Biophys. Res. Commun. 52 (2): 504–510. doi:10.1016/0006-291x(73)90740-7. PMID 4711166.
- ↑ Klug; Richmond (1984). "Structure of the nucleosome core particle at 7 Å resolution". Nature 311 (5986): 532–537. doi:10.1038/311532a0. PMID 6482966. Bibcode: 1984Natur.311..532R.
- ↑ Arents; Burlingame (1991). "The nucleosomal core histone octamer at 3.1 ˚A resolution: a tripartite protein assembly and a left-handed superhelix". PNAS 88 (22): 10148–52. doi:10.1073/pnas.88.22.10148. PMID 1946434. Bibcode: 1991PNAS...8810148A.
- ↑ Davey, Curt A.; Sargent, David F.; Luger, Karolin; Maeder, Armin W.; Richmond, Timothy J. (June 2002). "Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9Å Resolution". Journal of Molecular Biology 319 (5): 1097–1113. doi:10.1016/S0022-2836(02)00386-8. PMID 12079350.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 School, James D. Watson, Cold Spring Harbor Laboratory, Tania A. Baker, Massachusetts Institute of Technology, Stephen P. Bell, Massachusetts Institute of Technology, Alexander Gann, Cold Spring Harbor Laboratory, Michael Levine, University of California, Berkeley, Richard Losik, Harvard University ; with Stephen C. Harrison, Harvard Medical (2014). Molecular biology of the gene (Seventh ed.). Boston: Benjamin-Cummings Publishing Company. p. 241. ISBN 978-0321762436.
- ↑ 7.0 7.1 Luger, Karolin (April 2003). "Structure and dynamic behavior of nucleosomes". Current Opinion in Genetics & Development 13 (2): 127–135. doi:10.1016/S0959-437X(03)00026-1. PMID 12672489.
- ↑ D’Arcy, Sheena; Martin, Kyle W.; Panchenko, Tanya; Chen, Xu; Bergeron, Serge; Stargell, Laurie A.; Black, Ben E.; Luger, Karolin (September 2013). "Chaperone Nap1 Shields Histone Surfaces Used in a Nucleosome and Can Put H2A-H2B in an Unconventional Tetrameric Form". Molecular Cell 51 (5): 662–677. doi:10.1016/j.molcel.2013.07.015. PMID 23973327.
- ↑ Harshman, S. W.; Young, N. L.; Parthun, M. R.; Freitas, M. A. (14 August 2013). "H1 histones: current perspectives and challenges". Nucleic Acids Research 41 (21): 9593–9609. doi:10.1093/nar/gkt700. PMID 23945933.
- ↑ 10.0 10.1 Andrews, Andrew J.; Luger, Karolin (9 June 2011). "Nucleosome Structure(s) and Stability: Variations on a Theme". Annual Review of Biophysics 40 (1): 99–117. doi:10.1146/annurev-biophys-042910-155329. PMID 21332355.
- ↑ 11.0 11.1 11.2 Richmond, Timothy J.; Luger, Karolin; Mäder, Armin W.; Richmond, Robin K.; Sargent, David F. (18 September 1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution". Nature 389 (6648): 251–260. doi:10.1038/38444. PMID 9305837. Bibcode: 1997Natur.389..251L.
- ↑ 12.0 12.1 12.2 Biswas, Mithun; Voltz, Karine; Smith, Jeremy C.; Langowski, Jörg (15 December 2011). "Role of Histone Tails in Structural Stability of the Nucleosome". PLOS Computational Biology 7 (12): e1002279. doi:10.1371/journal.pcbi.1002279. PMID 22207822. Bibcode: 2011PLSCB...7E2279B.
- ↑ Becker, P. B. (16 September 2002). "NEW EMBO MEMBER'S REVIEW: Nucleosome sliding: facts and fiction". The EMBO Journal 21 (18): 4749–4753. doi:10.1093/emboj/cdf486. PMID 12234915.
- ↑ Fazzio, TG; Tsukiyama, T (November 2003). "Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism.". Molecular Cell 12 (5): 1333–40. doi:10.1016/s1097-2765(03)00436-2. PMID 14636590.
- ↑ Jenuwein, T; Allis, CD (Aug 10, 2001). "Translating the histone code". Science 293 (5532): 1074–80. doi:10.1126/science.1063127. PMID 11498575.
- ↑ Schiltz, RL; Mizzen, CA; Vassilev, A; Cook, RG; Allis, CD; Nakatani, Y (Jan 15, 1999). "Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates.". The Journal of Biological Chemistry 274 (3): 1189–92. doi:10.1074/jbc.274.3.1189. PMID 9880483.
- ↑ Jin, Q; Yu, LR; Wang, L; Zhang, Z; Kasper, LH; Lee, JE; Wang, C; Brindle, PK et al. (Jan 19, 2011). "Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation.". The EMBO Journal 30 (2): 249–62. doi:10.1038/emboj.2010.318. PMID 21131905.
- ↑ Yao, TP; Oh, SP; Fuchs, M; Zhou, ND; Ch'ng, LE; Newsome, D; Bronson, RT; Li, E et al. (May 1, 1998). "Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300.". Cell 93 (3): 361–72. doi:10.1016/S0092-8674(00)81165-4. PMID 9590171.
External links
 |
